New model for the yeast RNA polymerase I transcription cycle
- PMID: 11438642
- PMCID: PMC87188
- DOI: 10.1128/MCB.21.15.4847-4855.2001
New model for the yeast RNA polymerase I transcription cycle
Abstract
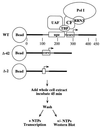
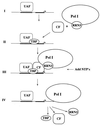
Using an immobilized template assay, we observed two steps in assembly of the yeast RNA polymerase I (Pol I) preinitiation complex: stable binding of upstream activating factor (UAF) followed by recruitment of Pol I-Rrn3p and core factor (CF). Pol I is required for stable association of CF with the promoter and can be recruited in the absence of Rrn3p. Upon transcription initiation, Pol I-Rrn3p and CF dissociate from the promoter while UAF remains behind. These findings support a novel model in which the Pol I basal machinery cycles on and off the promoter with each round of transcription. This model accounts for previous observations that rRNA synthesis may be controlled by regulating both promoter accessibility and polymerase activity.
Figures





References
-
- Carles C, Riva M. Yeast RNA polymerase I subunits and genes. In: Paule I, editor. Transcription of ribosomal RNA genes by eukaryotic RNA polymerase. Berlin, Germany: Springer Verlag; 1998.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
