Genome-wide gene expression profiles of the developing mouse hippocampus
- PMID: 11438693
- PMCID: PMC37526
- DOI: 10.1073/pnas.141244998
Genome-wide gene expression profiles of the developing mouse hippocampus
Abstract
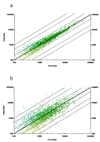
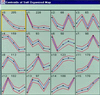
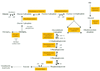
We have analyzed the developmental molecular programs of the mouse hippocampus, a cortical structure critical for learning and memory, by means of large-scale DNA microarray techniques. Of 11,000 genes and expressed sequence tags examined, 1,926 showed dynamic changes during hippocampal development from embryonic day 16 to postnatal day 30. Gene-cluster analysis was used to group these genes into 16 distinct clusters with striking patterns that appear to correlate with major developmental hallmarks and cellular events. These include genes involved in neuronal proliferation, differentiation, and synapse formation. A complete list of the transcriptional changes has been compiled into a comprehensive gene profile database (http://BrainGenomics.Princeton.edu), which should prove valuable in advancing our understanding of the molecular and genetic programs underlying both the development and the functions of the mammalian brain.
Figures

References
-
- Squire L R. Memory and Brain. New York: Oxford Univ. Press; 1997.
-
- O'keefe J, Nadel L. The Hippocampus as a Cognitive Map. Oxford: Clarendon; 1978.
-
- Cohen N J, Eichenbaum H. Memory, Amnesia, and Hippocampal System. Cambridge, MA: MIT Press; 1993.
-
- Bear M F, Malenka R L. Curr Opin Neurobiol. 1994;4:389–399. - PubMed
-
- Bliss T V P, Collingridge G L. Nature (London) 1993;361:31–39. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

