Functional dynamics in the active site of the ribonuclease binase
- PMID: 11438724
- PMCID: PMC35402
- DOI: 10.1073/pnas.121069998
Functional dynamics in the active site of the ribonuclease binase
Abstract
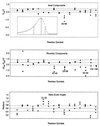
Binase, a member of a family of microbial guanyl-specific ribonucleases, catalyzes the endonucleotic cleavage of single-stranded RNA. It shares 82% amino acid identity with the well-studied protein barnase. We used NMR spectroscopy to study the millisecond dynamics of this small enzyme, using several methods including the measurement of residual dipolar couplings in solution. Our data show that the active site of binase is flanked by loops that are flexible at the 300-micros time scale. One of the catalytic residues, His-101, is located on such a flexible loop. In contrast, the other catalytic residue, Glu-72, is located on a beta-sheet, and is static. The residues Phe-55, part of the guanine base recognition site, and Tyr-102, stabilizing the base, are the most dynamic. Our findings suggest that binase possesses an active site that has a well-defined bottom, but which has sides that are flexible to facilitate substrate access/egress, and to deliver one of the catalytic residues. The motion in these loops does not change on complexation with the inhibitor d(CGAG) and compares well with the maximum k(cat) (1,500 s(-1)) of these ribonucleases. This observation indicates that the NMR-measured loop motions reflect the opening necessary for product release, which is apparently rate limiting for the overall turnover.
Figures


References
-
- Fersht A. Structure and Mechanism in Protein Science. New York: Freeman; 1999. p. 51.
-
- Ishima R, Torchia D A. Nat Struct Biol. 2000;7:740–743. - PubMed
-
- Yakovlev G I, Moiseyev G P, Struminskaya N K, Borzykh O A, Kipenskaya L V, Znamenskaya L V, Leschinskaya I B, Chernokalskaya E B, Hartley R W. FEBS Lett. 1994;354:305–306. - PubMed
-
- Meiering E M, Serrano L, Fersht A R. J Mol Biol. 1992;225:585–589. - PubMed
-
- Carrington A, McLachlan A. Introduction to Magnetic Resonance with Applications to Chemistry and Chemical Physics. New York: Harper & Row; 1967.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

