Genome size determination and coding capacity of Sodalis glossinidius, an enteric symbiont of tsetse flies, as revealed by hybridization to Escherichia coli gene arrays
- PMID: 11443086
- PMCID: PMC95346
- DOI: 10.1128/JB.183.15.4517-4525.2001
Genome size determination and coding capacity of Sodalis glossinidius, an enteric symbiont of tsetse flies, as revealed by hybridization to Escherichia coli gene arrays
Abstract
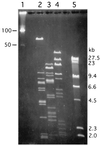
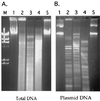
Recent molecular characterization of various microbial genomes has revealed differences in genome size and coding capacity between obligate symbionts and intracellular pathogens versus free-living organisms. Multiple symbiotic microorganisms have evolved with tsetse fly, the vector of African trypanosomes, over long evolutionary times. Although these symbionts are indispensable for tsetse fecundity, the biochemical and molecular basis of their functional significance is unknown. Here, we report on the genomic aspects of the secondary symbiont Sodalis glossinidius. The genome size of Sodalis is approximately 2 Mb. Its DNA is subject to extensive methylation and based on some of its conserved gene sequences has an A+T content of only 45%, compared to the typically AT-rich genomes of endosymbionts. Sodalis also harbors an extrachromosomal plasmid about 134 kb in size. We used a novel approach to gain insight into Sodalis genomic contents, i.e., hybridizing its DNA to macroarrays developed for Escherichia coli, a closely related enteric bacterium. In this analysis we detected 1,800 orthologous genes, corresponding to about 85% of the Sodalis genome. The Sodalis genome has apparently retained its genes for DNA replication, transcription, translation, transport, and the biosynthesis of amino acids, nucleic acids, vitamins, and cofactors. However, many genes involved in energy metabolism and carbon compound assimilation are apparently missing, which may indicate an adaptation to the energy sources available in the only nutrient of the tsetse host, blood. We present gene arrays as a rapid tool for comparative genomics in the absence of whole genome sequence to advance our understanding of closely related bacteria.
Figures






References
-
- Aksoy S. Tsetse: a haven for microorganisms. Parasitol Today. 2000;16:114–119. - PubMed
-
- Aksoy S. Wigglesworthia gen. nov. and Wigglesworthia glossinidia sp. nov., taxa consisting of the mycetocyte-associated, primary endosymbionts of tsetse flies. Int J Syst Bacteriol. 1995;45:848–851. - PubMed
-
- Aksoy S, Chen X, Hypsa V. Phylogeny and potential transmission routes of midgut-associated endosymbionts of tsetse (Diptera:Glossinidae) Insect Mol Biol. 1997;6:183–190. - PubMed
-
- Aksoy S, Pourhosseini A A, Chow A. Mycetome endosymbionts of tsetse flies constitute a distinct lineage related to Enterobacteriaceae. Insect Mol Biol. 1995;4:15–22. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

