DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide
- PMID: 11443091
- PMCID: PMC95351
- DOI: 10.1128/JB.183.15.4562-4570.2001
DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide
Abstract
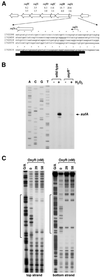
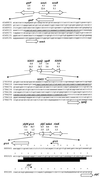
The genome-wide transcription profile of Escherichia coli cells treated with hydrogen peroxide was examined with a DNA microarray composed of 4,169 E. coli open reading frames. By measuring gene expression in isogenic wild-type and oxyR deletion strains, we confirmed that the peroxide response regulator OxyR activates most of the highly hydrogen peroxide-inducible genes. The DNA microarray measurements allowed the identification of several new OxyR-activated genes, including the hemH heme biosynthetic gene; the six-gene suf operon, which may participate in Fe-S cluster assembly or repair; and four genes of unknown function. We also identified several genes, including uxuA, encoding mannonate hydrolase, whose expression might be repressed by OxyR, since their expression was elevated in the DeltaoxyR mutant strain. In addition, the induction of some genes was found to be OxyR independent, indicating the existence of other peroxide sensors and regulators in E. coli. For example, the isc operon, which specifies Fe-S cluster formation and repair activities, was induced by hydrogen peroxide in strains lacking either OxyR or the superoxide response regulators SoxRS. These results expand our understanding of the oxidative stress response and raise interesting questions regarding the nature of other regulators that modulate gene expression in response to hydrogen peroxide.
Figures



References
-
- Bachmann B J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: ASM Press; 1996. pp. 2460–2488.
-
- Beale S I. Biosynthesis of hemes. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: ASM Press; 1996. pp. 731–748.
-
- Belkin S, Smulski D R, Dadon S, Vollmer A C, Van Dyk T K, LaRossa R A. A panel of stress-responsive luminous bacteria for the detection of selected classes of toxicants. Water Res. 1997;31:3009–3016.
-
- Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

