Computation-directed identification of OxyR DNA binding sites in Escherichia coli
- PMID: 11443092
- PMCID: PMC95352
- DOI: 10.1128/JB.183.15.4571-4579.2001
Computation-directed identification of OxyR DNA binding sites in Escherichia coli
Abstract
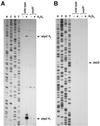
A computational search was carried out to identify additional targets for the Escherichia coli OxyR transcription factor. This approach predicted OxyR binding sites upstream of dsbG, encoding a periplasmic disulfide bond chaperone-isomerase; upstream of fhuF, encoding a protein required for iron uptake; and within yfdI. DNase I footprinting assays confirmed that oxidized OxyR bound to the predicted site centered 54 bp upstream of the dsbG gene and 238 bp upstream of a known OxyR binding site in the promoter region of the divergently transcribed ahpC gene. Although the new binding site was near dsbG, Northern blotting and primer extension assays showed that OxyR binding to the dsbG-proximal site led to the induction of a second ahpCF transcript, while OxyR binding to the ahpCF-proximal site leads to the induction of both dsbG and ahpC transcripts. Oxidized OxyR binding to the predicted site centered 40 bp upstream of the fhuF gene was confirmed by DNase I footprinting, but these assays further revealed a second higher-affinity site in the fhuF promoter. Interestingly, the two OxyR sites in the fhuF promoter overlapped with two regions bound by the Fur repressor. Expression analysis revealed that fhuF was repressed by hydrogen peroxide in an OxyR-dependent manner. Finally, DNase I footprinting experiments showed OxyR binding to the site predicted to be within the coding sequence of yfdI. These results demonstrate the versatile modes of regulation by OxyR and illustrate the need to learn more about the ensembles of binding sites and transcripts in the E. coli genome.
Figures






Similar articles
-
Molecular and physiological analysis of an OxyR-regulated ahpC promoter in Xanthomonas campestris pv. phaseoli.Mol Microbiol. 2000 Sep;37(6):1504-14. doi: 10.1046/j.1365-2958.2000.02107.x. Mol Microbiol. 2000. PMID: 10998180
-
Interactions of OxyR with the promoter region of the oxyR and ahpC genes from Mycobacterium leprae and Mycobacterium tuberculosis.J Bacteriol. 1997 Apr;179(7):2401-9. doi: 10.1128/jb.179.7.2401-2409.1997. J Bacteriol. 1997. PMID: 9079928 Free PMC article.
-
Identification and molecular analysis of oxyR-regulated promoters important for the bacterial adaptation to oxidative stress.J Mol Biol. 1989 Dec 20;210(4):709-19. doi: 10.1016/0022-2836(89)90104-6. J Mol Biol. 1989. PMID: 2693740
-
Members of the Fur protein family regulate iron and zinc transport in E. coli and characteristics of the Fur-regulated fhuF protein.J Mol Microbiol Biotechnol. 2002 May;4(3):217-22. J Mol Microbiol Biotechnol. 2002. PMID: 11931550 Review.
-
The OxyR regulon.Antonie Van Leeuwenhoek. 1990 Oct;58(3):157-61. doi: 10.1007/BF00548927. Antonie Van Leeuwenhoek. 1990. PMID: 2256675 Review.
Cited by
-
During Oxidative Stress the Clp Proteins of Escherichia coli Ensure that Iron Pools Remain Sufficient To Reactivate Oxidized Metalloenzymes.J Bacteriol. 2020 Aug 25;202(18):e00235-20. doi: 10.1128/JB.00235-20. Print 2020 Aug 25. J Bacteriol. 2020. PMID: 32601069 Free PMC article.
-
OxyR-regulated T6SS functions in coordination with siderophore to resist oxidative stress.Microbiol Spectr. 2024 Feb 6;12(2):e0323123. doi: 10.1128/spectrum.03231-23. Epub 2024 Jan 8. Microbiol Spectr. 2024. PMID: 38189330 Free PMC article.
-
Transcriptional regulation mechanism of ter operon by OxyR in Yersinia pestis.Curr Microbiol. 2014 Jul;69(1):42-6. doi: 10.1007/s00284-014-0550-7. Epub 2014 Mar 1. Curr Microbiol. 2014. PMID: 24577613
-
The OxyR-regulated phnW gene encoding 2-aminoethylphosphonate:pyruvate aminotransferase helps protect Pseudomonas aeruginosa from tert-butyl hydroperoxide.PLoS One. 2017 Dec 7;12(12):e0189066. doi: 10.1371/journal.pone.0189066. eCollection 2017. PLoS One. 2017. PMID: 29216242 Free PMC article.
-
PerR functions as a redox-sensing transcription factor regulating metal homeostasis in the thermoacidophilic archaeon Saccharolobus islandicus REY15A.Nucleic Acids Res. 2025 Jan 7;53(1):gkae1263. doi: 10.1093/nar/gkae1263. Nucleic Acids Res. 2025. PMID: 39727184 Free PMC article.
References
-
- Andersen C L, Matthey-Dupraz A, Missiakas D, Raina S. A new Escherichia coli gene, dsbG, encodes a periplasmic protein involved in disulphide bond formation, required for recycling DsbA/DsbB and DsbC redox proteins. Mol Microbiol. 1997;26:121–132. - PubMed
-
- Bessette P H, Cotto J J, Gilbert H F, Georgiou G. In vivo and in vitro function of the Escherichia coli periplasmic cysteine oxidoreductase DsbG. J Biol Chem. 1999;274:7784–7792. - PubMed
-
- Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. - PubMed
-
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. - PubMed
-
- Debarbieux L, Beckwith J. Electron avenue: pathways of disulfide bond formation and isomerization. Cell. 1999;99:117–119. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

