Activation by gene amplification of pitB, encoding a third phosphate transporter of Escherichia coli K-12
- PMID: 11443103
- PMCID: PMC95363
- DOI: 10.1128/JB.183.15.4659-4663.2001
Activation by gene amplification of pitB, encoding a third phosphate transporter of Escherichia coli K-12
Abstract
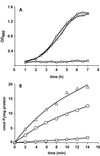
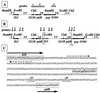
Two systems for the uptake of inorganic phosphate (P(i)) in Escherichia coli, PitA and Pst, have been described. A revertant of a pitA pstS double mutant that could grow on P(i) was isolated. We demonstrate that the expression of a new P(i) transporter, PitB, is activated in this strain by a gene amplification event.
Figures

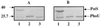
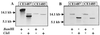

Similar articles
-
The phosphate-binding protein of Escherichia coli is not essential for P(i)-regulated expression of the pho regulon.J Bacteriol. 2001 Oct;183(19):5768-71. doi: 10.1128/JB.183.19.5768-5771.2001. J Bacteriol. 2001. PMID: 11544243 Free PMC article.
-
Characterization of PitA and PitB from Escherichia coli.J Bacteriol. 2001 Sep;183(17):5008-14. doi: 10.1128/JB.183.17.5008-5014.2001. J Bacteriol. 2001. PMID: 11489853 Free PMC article.
-
The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the pho regulon.J Bacteriol. 1997 Apr;179(8):2534-9. doi: 10.1128/jb.179.8.2534-2539.1997. J Bacteriol. 1997. PMID: 9098050 Free PMC article.
-
Molybdate transport.Res Microbiol. 2001 Apr-May;152(3-4):311-21. doi: 10.1016/s0923-2508(01)01202-5. Res Microbiol. 2001. PMID: 11421278 Review.
-
More than just "histone-like" proteins.Cell. 1990 Nov 2;63(3):451-3. doi: 10.1016/0092-8674(90)90438-k. Cell. 1990. PMID: 2121364 Review. No abstract available.
Cited by
-
Isolation, Characterization, and Tea Growth-Promoting Analysis of JW-CZ2, a Bacterium With 1-Aminocyclopropane-1-Carboxylic Acid Deaminase Activity Isolated From the Rhizosphere Soils of Tea Plants.Front Microbiol. 2022 Feb 28;13:792876. doi: 10.3389/fmicb.2022.792876. eCollection 2022. Front Microbiol. 2022. PMID: 35295310 Free PMC article.
-
Induction of the Pho regulon suppresses the growth defect of an Escherichia coli sgrS mutant, connecting phosphate metabolism to the glucose-phosphate stress response.J Bacteriol. 2012 May;194(10):2520-30. doi: 10.1128/JB.00009-12. Epub 2012 Mar 16. J Bacteriol. 2012. PMID: 22427626 Free PMC article.
-
The phosphate starvation stimulon of Corynebacterium glutamicum determined by DNA microarray analyses.J Bacteriol. 2003 Aug;185(15):4519-29. doi: 10.1128/JB.185.15.4519-4529.2003. J Bacteriol. 2003. PMID: 12867461 Free PMC article.
-
Autoamplification of a two-component regulatory system results in "learning" behavior.J Bacteriol. 2001 Aug;183(16):4914-7. doi: 10.1128/JB.183.16.4914-4917.2001. J Bacteriol. 2001. PMID: 11466297 Free PMC article.
-
Strategies of organic phosphorus recycling by soil bacteria: acquisition, metabolism, and regulation.Environ Microbiol Rep. 2022 Feb;14(1):3-24. doi: 10.1111/1758-2229.13040. Epub 2022 Jan 10. Environ Microbiol Rep. 2022. PMID: 35001516 Free PMC article. Review.
References
-
- Agterberg M, Fransen R, Tommassen J. Expression of Escherichia coli PhoE protein in avirulent Salmonella typhimurium aroA and galE strains. FEMS Microbiol Lett. 1988;50:295–299.
-
- Alexeyev M F, Shokolenko I N, Croughan T P. Improved antibiotic-resistance gene cassettes and omega elements for Escherichia coli vector construction and in vitro deletion/insertion mutagenesis. Gene. 1995;160:63–67. - PubMed
-
- Bennett R L, Malamy M H. Arsenate resistant mutants of Escherichia coli and phosphate transport. Biochem Biophys Res Commun. 1970;40:496–503. - PubMed
-
- Blattner F R, Plunkett III G, Block C A, Perna N T, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. - PubMed
-
- Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–544. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

