Global analysis of ATM polymorphism reveals significant functional constraint
- PMID: 11443540
- PMCID: PMC1235311
- DOI: 10.1086/321296
Global analysis of ATM polymorphism reveals significant functional constraint
Abstract
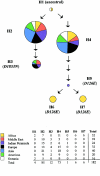
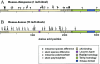
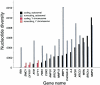
ATM, the gene that is mutated in ataxia-telangiectasia, is associated with cerebellar degeneration, abnormal proliferation of small blood vessels, and cancer. These clinically important manifestations have stimulated interest in defining the sequence variation in the ATM gene. Therefore, we undertook a comprehensive survey of sequence variation in ATM in diverse human populations. The protein-encoding exons of the gene (9,168 bp) and the adjacent intron and untranslated sequences (14,661 bp) were analyzed in 93 individuals from seven major human populations. In addition, the coding sequence was analyzed in one chimpanzee, one gorilla, one orangutan, and one Old World monkey. In human ATM, 88 variant sites were discovered by denaturing high-performance liquid chromatography, which is 96%-100% sensitive for detection of DNA sequence variation. ATM was compared to 14 other autosomal genes for nucleotide diversity. The noncoding regions of ATM had diversity values comparable to other genes, but the coding regions had very low diversity, especially in the last 29% of the protein sequence. A test of the neutral evolution hypothesis, through use of the Hudson/Kreitman/Aguadé statistic, revealed that this region of the human ATM gene was significantly constrained relative to that of the orangutan, the Old World monkey, and the mouse, but not relative to that of the chimpanzee or the gorilla. ATM displayed extensive linkage disequilibrium, consistent with suppression of meiotic recombination at this locus. Seven haplotypes were defined. Two haplotypes accounted for 82% of all chromosomes analyzed in all major populations; two others carrying the same D126E missense polymorphism accounted for 33% of chromosomes in Africa but were never observed outside of Africa. The high frequency of this polymorphism may be due either to a population expansion within Africa or to selective pressure.
Figures

Comment in
-
Resolving ATM haplotypes in whites.Am J Hum Genet. 2003 Apr;72(4):1071-3. doi: 10.1086/373879. Am J Hum Genet. 2003. PMID: 12708462 Free PMC article. No abstract available.
References
Electronic-Database Information
-
- Berkeley Drosophila Genome Project, http://www.fruitfly.org/seq_tools/splice.html (for splice site prediction algorithm)
-
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/ (for polymorphisms for genes)
-
- DNA Variation Group, http://insertion.stanford.edu/melt.html (for DHPLC Melt program for prediction of DHPLC analysis temperature)
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for genomic sequences of ATM [accession number HSU82828]; ABCB1 [accession number AC002457]; AMPD1 [accession number M60092]; BRCA1 [accession number U14680]; BRCA2 [accession number U43746]; CACNA1A [accession number AF004884]; COX2 [accession number U04636]; FBN1 [accession number L13923]; IL4 [accession number M23442]; MMP1, MMP3, and MMP12 [accession number U78045]; RB1 [accession number M15400]; WRN [accession number AF091214]; XRCC1 [accession number M36089]; and for the mRNA and amino acid sequences of mouse Atm [accession number MMU43678])
-
- LocusLink, http://www.ncbi.nlm.nih.gov/LocusLink/ (for gene sequences)
References
-
- Bay J, Grancho M, Pernin D, Presneau N, Rio P, Tchirkov A, Uhrhammer N, Verrelle P, Gatti R, Bignon Y (1998) No evidence for constitutional ATM mutation in breast/gastric cancer families. Int J Oncol 12:1385–1390 - PubMed
-
- Bay J, Uhrhammer N, Pernin D, Presneau N, Tchirkov A, Vuillaume M, Laplace V, Grancho M, Verrelle P, Hall J, Bignon Y (1999) High incidence of cancer in a family segregating a mutation of the ATM gene: possible role of ATM heterozygosity in cancer. Hum Mutat 14:485–492 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

