Crystal structure of the M-fragment of alpha-catenin: implications for modulation of cell adhesion
- PMID: 11447106
- PMCID: PMC125534
- DOI: 10.1093/emboj/20.14.3645
Crystal structure of the M-fragment of alpha-catenin: implications for modulation of cell adhesion
Abstract
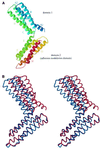
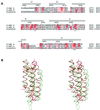
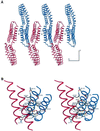
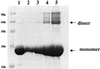
The cytoskeletal protein alpha-catenin, which shares structural similarity with vinculin, is required for cadherin-mediated cell adhesion, and functions to modulate cell adhesive strength and to link the cadherins to the actin-based cytoskeleton. Here we describe the crystal structure of a region of alpha-catenin (residues 377-633) termed the M-fragment. The M-fragment is composed of a tandem repeat of two antiparallel four-helix bundles of virtually identical architectures that are related in structure to the dimerization domain of alpha-catenin and the tail region of vinculin. These results suggest that alpha-catenin is composed of repeating antiparallel helical domains. The region of alpha-catenin previously defined as an adhesion modulation domain corresponds to the C-terminal four-helix bundle of the M-fragment, and in the crystal lattice these domains exist as dimers. Evidence for dimerization of the M-fragment of alpha-catenin in solution was detected by chemical cross-linking experiments. The tendency of the adhesion modulation domain to form dimers may explain its biological activity of promoting cell-cell adhesiveness by inducing lateral dimerization of the associated cadherin molecule.
Figures



References
-
- Aberle H., Butz,S., Stappert,J., Weissig,H., Kemler,R. and Hoschuetzky,H. (1994) Assembly of the cadherin–catenin complex in vitro with recombinant proteins. J. Cell Sci., 107, 3655–3663. - PubMed
-
- Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determination of the bovine mitochondrial F1 ATPase. Acta Crystallogr. D, 52, 30–42. - PubMed
-
- Bakolitsa C., de Pereda,J.M., Bagshaw,C.R., Critchley,D.R. and Liddington,R.C. (1999) Crystal structure of the vinculin tail suggests a pathway for activation. Cell, 99, 603–613. - PubMed
-
- Birchmeier W. and Behrens,J. (1994) Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochim. Biophys. Acta, 1198, 11–26. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

