Synthesis of a novel hepatitis C virus protein by ribosomal frameshift
- PMID: 11447125
- PMCID: PMC125543
- DOI: 10.1093/emboj/20.14.3840
Synthesis of a novel hepatitis C virus protein by ribosomal frameshift
Abstract
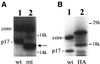
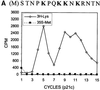
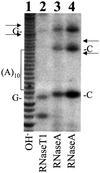
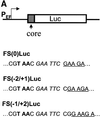
Hepatitis C virus (HCV) is an important human pathogen that affects approximately 100 million people worldwide. Its RNA genome codes for a polyprotein, which is cleaved by viral and cellular proteases to produce at least 10 mature viral protein products. We report here the discovery of a novel HCV protein synthesized by ribosomal frameshift. This protein, which we named the F protein, is synthesized from the initiation codon of the polyprotein sequence followed by ribosomal frameshift into the -2/+1 reading frame. This ribosomal frameshift requires only codons 8-14 of the core protein-coding sequence, and the shift junction is located at or near codon 11. An F protein analog synthesized in vitro reacted with the sera of HCV patients but not with the sera of hepatitis B patients, indicating the expression of the F protein during natural HCV infection. This unexpected finding may open new avenues for the development of anti-HCV drugs.
Figures








References
-
- Beard M.R. et al. (1999) An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology, 30, 316–324. - PubMed
-
- Blight K.J., Kolykhalov,A.A. and Rice,C.M. (2000) Efficient initiation of HCV RNA replication in cell culture. Science, 290, 1972–1974. - PubMed
-
- Bukh J., Apgar,C.L. and Yanagi,M. (1999) Toward a surrogate model for hepatitis C virus: an infectious molecular clone of the GB virus-B hepatitis agent. Virology, 262, 470–478. - PubMed
-
- Choo Q.-L., Kuo,G., Weiner,A.J., Overby,L.R., Bradley,D.W. and Houghton,M. (1989) Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science, 244, 359–362. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

