Two overlapping reading frames in a single exon encode interacting proteins--a novel way of gene usage
- PMID: 11447126
- PMCID: PMC125537
- DOI: 10.1093/emboj/20.14.3849
Two overlapping reading frames in a single exon encode interacting proteins--a novel way of gene usage
Abstract
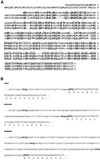
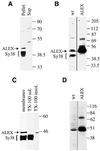
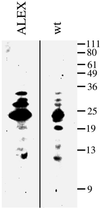
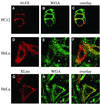
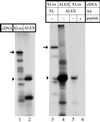
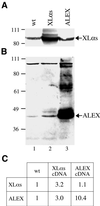
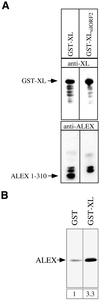
The >1 kb XL-exon of the rat XLalphas/Galphas gene encodes the 37 kDa XL-domain, the N-terminal half of the 78 kDa neuroendocrine-specific G-protein alpha-subunit XLalphas. Here, we describe a novel feature of the XL-exon, the presence of an alternative >1 kb open reading frame (ORF) that completely overlaps with the ORF encoding the XL-domain. The alternative ORF starts 32 nucleotides downstream of the start codon for the XL-domain and is terminated by a stop codon exactly at the end of the XL-exon. The alternative ORF encodes ALEX, a very basic (pI 11.8), proline-rich protein of 356 amino acids. Both XLalphas and ALEX are translated from the same mRNA. Like XLalphas, ALEX is expressed in neuroendocrine cells and tightly associated with the cytoplasmic leaflet of the plasma membrane. Remarkably, ALEX binds to the XL-domain of XLalphas. Our results reveal a mechanism of gene usage that is without precedent in mammalian genomes.
Figures




References
-
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds.) (1997) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY.
-
- Barrell B.G., Air,G.M. and Hutchinson,C.A. (1976) Overlapping genes in bacteriophage φX174. Nature, 264, 34–41. - PubMed
-
- Barrell B.G., Shaw,D.C., Walker,J.E., Northop,F.D., Godson,G.N. and Fiddes,J.C. (1978) Overlapping genes in bacteriophages φX174 and G4. Biochem. Soc. Trans., 6, 63–67. - PubMed
-
- Bartsch D., Casadio,A., Karl,K.A., Serodio,P. and Kandel,E.R. (1998) CREB1 encodes a nuclear activator, a repressor and a cytoplasmatic modulator that form a regulatory unit critical for long-term facilitation. Cell, 95, 211–223. - PubMed
-
- Cleveland, D.W., Fischer S.G., Kirschner M.W. and Laemmli U.K. (1977) Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J. Biol. Chem., 252, 1102–1106. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

