Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells
- PMID: 11448991
- PMCID: PMC2196857
- DOI: 10.1083/jcb.200101089
Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells
Abstract
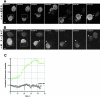
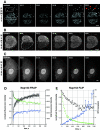
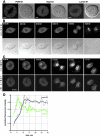
The nuclear pore complex (NPC) and its relationship to the nuclear envelope (NE) was characterized in living cells using POM121-green fluorescent protein (GFP) and GFP-Nup153, and GFP-lamin B1. No independent movement of single pore complexes was found within the plane of the NE in interphase. Only large arrays of NPCs moved slowly and synchronously during global changes in nuclear shape, strongly suggesting mechanical connections which form an NPC network. The nuclear lamina exhibited identical movements. NPC turnover measured by fluorescence recovery after photobleaching of POM121 was less than once per cell cycle. Nup153 association with NPCs was dynamic and turnover of this nucleoporin was three orders of magnitude faster. Overexpression of both nucleoporins induced the formation of annulate lamellae (AL) in the endoplasmic reticulum (ER). Turnover of AL pore complexes was much higher than in the NE (once every 2.5 min). During mitosis, POM121 and Nup153 were completely dispersed and mobile in the ER (POM121) or cytosol (Nup153) in metaphase, and rapidly redistributed to an immobilized pool around chromatin in late anaphase. Assembly and immobilization of both nucleoporins occurred before detectable recruitment of lamin B1, which is thus unlikely to mediate initiation of NPC assembly at the end of mitosis.
Figures




Comment in
-
Nuclear pore complexes: dynamics in unexpected places.J Cell Biol. 2001 Jul 9;154(1):17-20. doi: 10.1083/jcb.200106071. J Cell Biol. 2001. PMID: 11448985 Free PMC article. Review.
References
-
- Allen, T.D., J.M. Cronshaw, S. Bagley, E. Kiseleva, and M.W. Goldberg. 2000. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J. Cell Sci. 113:1651–1659. - PubMed
-
- Belgareh, N., and V. Doye. 1999. Yeast and vertebrate nuclear pore complexes: evolutionary conserved, yet divergent macromolecular assemblies. Protoplasma. 209:133–143.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

