Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants
- PMID: 11449047
- PMCID: PMC139543
- DOI: 10.1105/tpc.010011
Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants
Abstract
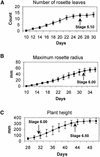
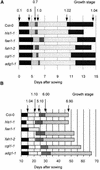
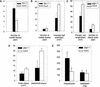
With the completion of the Arabidopsis genome sequencing project, the next major challenge is the large-scale determination of gene function. As a model organism for agricultural biotechnology, Arabidopsis presents the opportunity to provide key insights into the way that gene function can affect commercial crop production. In an attempt to aid in the rapid discovery of gene function, we have established a high throughput phenotypic analysis process based on a series of defined growth stages that serve both as developmental landmarks and as triggers for the collection of morphological data. The data collection process has been divided into two complementary platforms to ensure the capture of detailed data describing Arabidopsis growth and development over the entire life of the plant. The first platform characterizes early seedling growth on vertical plates for a period of 2 weeks. The second platform consists of an extensive set of measurements from plants grown on soil for a period of approximately 2 months. When combined with parallel processes for metabolic and gene expression profiling, these platforms constitute a core technology in the high throughput determination of gene function. We present here analyses of the development of wild-type Columbia (Col-0) plants and selected mutants to illustrate a framework methodology that can be used to identify and interpret phenotypic differences in plants resulting from genetic variation and/or environmental stress.
Figures


References
-
- Hartenstein, V. (1993). Atlas of Drosophila Morphology and Development. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

