Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240-->Gly
- PMID: 11451684
- PMCID: PMC90641
- DOI: 10.1128/AAC.45.8.2269-2275.2001
Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240-->Gly
Abstract
Three clinical strains (Escherichia coli Rio-6, E. coli Rio-7, and Enterobacter cloacae Rio-9) collected in 1996 and 1999 from hospitals in Rio de Janeiro (Brazil) were resistant to broad-spectrum cephalosporins and gave a positive double-disk synergy test. Two bla(CTX-M) genes encoding beta-lactamases of pl 7.9 and 8.2 were implicated in this resistance: the bla(CTX-M-9) gene observed in E. coli Rio-7 and E. cloacae Rio-9 and a novel CTX-M-encoding gene, designated bla(CTX-M-16), observed in E. coli strain Rio-6. The deduced amino acid sequence of CTX-M-16 differed from CTX-M-9 only by the substitution Asp-240-->Gly. The CTX-M-16-producing E. coli transformant exhibited the same level of resistance to cefotaxime (MIC, 16 microg/ml) but had a higher MIC of ceftazidime (MIC, 8 versus 1 microg/ml) than the CTX-M-9-producing transformant. Enzymatic studies revealed that CTX-M-16 had a 13-fold higher affinity for aztreonam and a 7.5-fold higher k(cat) for ceftazidime than CTX-M-9, thereby showing that the residue in position 240 can modulate the enzymatic properties of CTX-M enzymes. The two bla(CTX-M-9) genes and the bla(CTX-M-16) gene were located on different plasmids, suggesting the presence of mobile elements associated with CTX-M-encoding genes. CTX-M-2 and CTX-M-8 enzymes were found in Brazil in 1996, and two other CTX-M beta-lactamases, CTX-M-9 and CTX-M-16, were subsequently observed. These reports are evidence of the diversity of CTX-M-type extended-spectrum beta-lactamases in Brazil.
Figures
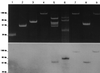

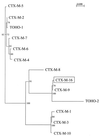

Comment in
-
Early dissemination of CTX-M-derived enzymes in South America.Antimicrob Agents Chemother. 2002 Feb;46(2):602-4. doi: 10.1128/AAC.46.2.602-604.2002. Antimicrob Agents Chemother. 2002. PMID: 11796390 Free PMC article. No abstract available.
References
-
- Barthélémy M, Péduzzi J, Bernard H, Tancrede C, Labia R. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum β-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim Biophys Acta. 1992;1122:15–22. - PubMed
-
- Bauernfeind A, Casellas J M, Goldberg M, Holley M, Jungwirth R, Mangold P, Rohnisch T, Schweighart S, Wilhelm R. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection. 1992;20:158–163. - PubMed
-
- Bauernfeind A, Grimm H, Schweighart S. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection. 1990;18:294–298. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

