Macrolide resistance gene mreA of Streptococcus agalactiae encodes a flavokinase
- PMID: 11451686
- PMCID: PMC90643
- DOI: 10.1128/AAC.45.8.2280-2286.2001
Macrolide resistance gene mreA of Streptococcus agalactiae encodes a flavokinase
Abstract
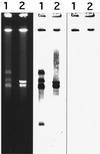
The mreA gene from Streptococcus agalactiae COH31 gamma/delta, resistant to macrolides and clindamycin by active efflux, has recently been cloned in Escherichia coli, where it was reported to confer macrolide resistance (J. Clancy, F. Dib-Hajj, J. W. Petitpas, and W. Yuan, Antimicrob. Agents Chemother. 41:2719--2723, 1997). Cumulative data suggested that the mreA gene was located on the chromosome of S. agalactiae COH31 gamma/delta. Analysis of the deduced amino acid sequence of mreA revealed significant homology with several bifunctional flavokinases/(flavin adenine dinucleotide (FAD) synthetases, which convert riboflavin to flavin mononucleotide (FMN) and FMN to FAD, respectively. High-performance liquid chromatography experiments showed that the mreA gene product had a monofunctional flavokinase activity, similar to that of RibR from Bacillus subtilis. Sequences identical to those of the mreA gene and of a 121-bp upstream region containing a putative promoter were detected in strains of S. agalactiae UCN4, UCN5, and UCN6 susceptible to macrolides. mreA and its allele from S. agalactiae UCN4 were cloned on the shuttle vector pAT28. Both constructs were introduced into E. coli, where they conferred a similar two- to fourfold increase in the MICs of erythromycin, spiramycin, and clindamycin. The MICs of a variety of other molecules, including crystal violet, acriflavin, sodium dodecyl sulfate, and antibiotics, such as certain cephalosporins, chloramphenicol, doxycycline, nalidixic acid, novobiocin, and rifampin, were also increased. In contrast, resistance to these compounds was not detected when the constructs were introduced into E. faecalis JH2-2. In conclusion, the mreA gene was probably resident in S. agalactiae and may encode a metabolic function. We could not provide any evidence that it was responsible for macrolide resistance in S. agalactiae COH31 gamma/delta; broad-spectrum resistance conferred by the gene in E. coli could involve multidrug efflux pumps by a mechanism that remains to be elucidated.
Figures


Similar articles
-
Cloning and characterization of a novel macrolide efflux gene, mreA, from Streptococcus agalactiae.Antimicrob Agents Chemother. 1997 Dec;41(12):2719-23. doi: 10.1128/AAC.41.12.2719. Antimicrob Agents Chemother. 1997. PMID: 9420045 Free PMC article.
-
The ribR gene encodes a monofunctional riboflavin kinase which is involved in regulation of the Bacillus subtilis riboflavin operon.Microbiology (Reading). 1999 Jan;145 ( Pt 1):67-73. doi: 10.1099/13500872-145-1-67. Microbiology (Reading). 1999. PMID: 10206712
-
The bifunctional flavokinase/flavin adenine dinucleotide synthetase from Streptomyces davawensis produces inactive flavin cofactors and is not involved in resistance to the antibiotic roseoflavin.J Bacteriol. 2008 Mar;190(5):1546-53. doi: 10.1128/JB.01586-07. Epub 2007 Dec 21. J Bacteriol. 2008. PMID: 18156273 Free PMC article.
-
Resistance phenotypes conferred by macrolide phosphotransferases.FEMS Microbiol Lett. 2007 Apr;269(2):317-22. doi: 10.1111/j.1574-6968.2007.00643.x. Epub 2007 Feb 16. FEMS Microbiol Lett. 2007. PMID: 17302923
-
New lnu(C) gene conferring resistance to lincomycin by nucleotidylation in Streptococcus agalactiae UCN36.Antimicrob Agents Chemother. 2005 Jul;49(7):2716-9. doi: 10.1128/AAC.49.7.2716-2719.2005. Antimicrob Agents Chemother. 2005. PMID: 15980341 Free PMC article.
Cited by
-
Molecular characteristics and antibiotic resistance mechanisms of clindamycin-resistant Streptococcus agalactiae isolates in China.Front Microbiol. 2023 Mar 1;14:1138039. doi: 10.3389/fmicb.2023.1138039. eCollection 2023. Front Microbiol. 2023. PMID: 36937303 Free PMC article.
-
Dispersal history and bidirectional human-fish host switching of invasive, hypervirulent Streptococcus agalactiae sequence type 283.PLOS Glob Public Health. 2023 Oct 19;3(10):e0002454. doi: 10.1371/journal.pgph.0002454. eCollection 2023. PLOS Glob Public Health. 2023. PMID: 37856430 Free PMC article.
-
Genome analysis of Streptococcus spp. isolates from animals in pre-antibiotic era with respect to antibiotic susceptibility and virulence gene profiles.Vet Res. 2024 Apr 15;55(1):51. doi: 10.1186/s13567-024-01302-0. Vet Res. 2024. PMID: 38622639 Free PMC article.
-
Mining microbial metatranscriptomes for expression of antibiotic resistance genes under natural conditions.Sci Rep. 2015 Jul 8;5:11981. doi: 10.1038/srep11981. Sci Rep. 2015. PMID: 26153129 Free PMC article.
-
Bacteriocin Producing Streptococcus agalactiae Strains Isolated from Bovine Mastitis in Brazil.Microorganisms. 2022 Mar 9;10(3):588. doi: 10.3390/microorganisms10030588. Microorganisms. 2022. PMID: 35336163 Free PMC article.
References
-
- Bacher A. Riboflavin kinase and FAD synthetase. In: Müller F, editor. Chemistry and biochemistry of flavoenzymes. Vol. 1. Boca Raton, Fla: CRC Press; 1991. pp. 349–370.
-
- Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Clancy J, Petitpas J, Dib-Hajj F, Yuan W, Cronan M, Kamath A V, Bergeron J, Retsema J A. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol Microbiol. 1996;22:867–879. - PubMed
-
- Comité de l'Antibiogramme de la Société Française de Microbiologie. 1996 report of the Comité de l'Antibiogramme de la Société Française de Microbiologie. Technical recommendations for in vitro susceptibility testing. Clin Microbiol Infect. 1996;2S1:11–25.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

