A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae
- PMID: 11452010
- PMCID: PMC55669
- DOI: 10.1091/mbc.12.7.2147
A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae
Abstract
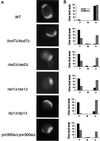
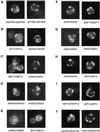
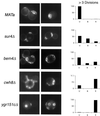
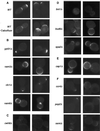
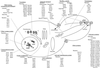
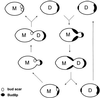
A genome-wide screen of 4168 homozygous diploid yeast deletion strains has been performed to identify nonessential genes that participate in the bipolar budding pattern. By examining bud scar patterns representing the sites of previous cell divisions, 127 mutants representing three different phenotypes were found: unipolar, axial-like, and random. From this screen, 11 functional classes of known genes were identified, including those involved in actin-cytoskeleton organization, general bud site selection, cell polarity, vesicular transport, cell wall synthesis, protein modification, transcription, nuclear function, translation, and other functions. Four characterized genes that were not known previously to participate in bud site selection were also found to be important for the haploid axial budding pattern. In addition to known genes, we found 22 novel genes (20 are designated BUD13-BUD32) important for bud site selection. Deletion of one resulted in unipolar budding exclusively from the proximal pole, suggesting that this gene plays an important role in diploid distal budding. Mutations in 20 other novel BUD genes produced a random budding phenotype and one produced an axial-like budding defect. Several of the novel Bud proteins were fused to green fluorescence protein; two proteins were found to localize to sites of polarized cell growth (i.e., the bud tip in small budded cells and the neck in cells undergoing cytokinesis), similar to that postulated for the bipolar signals and proteins that target cell division site tags to their proper location in the cell. Four others localized to the nucleus, suggesting that they play a role in gene expression. The bipolar distal marker Bud8 was localized in a number of mutants; many showed an altered Bud8-green fluorescence protein localization pattern. Through the genome-wide identification and analysis of different mutants involved in bipolar bud site selection, an integrated pathway for this process is presented in which proximal and distal bud site selection tags are synthesized and localized at their appropriate poles, thereby directing growth at those sites. Genome-wide screens of defined collections of mutants hold significant promise for dissecting many biological processes in yeast.
Figures


Similar articles
-
Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae.Mol Cell Biol. 2000 Jul;20(14):5235-47. doi: 10.1128/MCB.20.14.5235-5247.2000. Mol Cell Biol. 2000. PMID: 10866679 Free PMC article.
-
Bud8p and Bud9p, proteins that may mark the sites for bipolar budding in yeast.Mol Biol Cell. 2001 Aug;12(8):2497-518. doi: 10.1091/mbc.12.8.2497. Mol Biol Cell. 2001. PMID: 11514631 Free PMC article.
-
A role for the actin cytoskeleton of Saccharomyces cerevisiae in bipolar bud-site selection.J Cell Biol. 1997 Jan 13;136(1):111-23. doi: 10.1083/jcb.136.1.111. J Cell Biol. 1997. PMID: 9008707 Free PMC article.
-
The final cut: cell polarity meets cytokinesis at the bud neck in S. cerevisiae.Cell Mol Life Sci. 2016 Aug;73(16):3115-36. doi: 10.1007/s00018-016-2220-3. Epub 2016 Apr 16. Cell Mol Life Sci. 2016. PMID: 27085703 Free PMC article. Review.
-
Functional genomics in the study of yeast cell polarity: moving in the right direction.Philos Trans R Soc Lond B Biol Sci. 2013 Sep 23;368(1629):20130118. doi: 10.1098/rstb.2013.0118. Print 2013. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 24062589 Free PMC article. Review.
Cited by
-
New Insights Into Lignification via Network and Multi-Omics Analyses of Arogenate Dehydratase Knock-Out Mutants in Arabidopsis thaliana.Front Plant Sci. 2021 May 25;12:664250. doi: 10.3389/fpls.2021.664250. eCollection 2021. Front Plant Sci. 2021. PMID: 34113365 Free PMC article.
-
Regulation of cell polarity by interactions of Msb3 and Msb4 with Cdc42 and polarisome components.Mol Cell Biol. 2005 Oct;25(19):8567-80. doi: 10.1128/MCB.25.19.8567-8580.2005. Mol Cell Biol. 2005. PMID: 16166638 Free PMC article.
-
Analysis of the interaction between piD261/Bud32, an evolutionarily conserved protein kinase of Saccharomyces cerevisiae, and the Grx4 glutaredoxin.Biochem J. 2004 Jan 15;377(Pt 2):395-405. doi: 10.1042/BJ20030638. Biochem J. 2004. PMID: 14519092 Free PMC article.
-
Role for Arf3p in development of polarity, but not endocytosis, in Saccharomyces cerevisiae.Mol Biol Cell. 2003 Sep;14(9):3834-47. doi: 10.1091/mbc.e03-01-0013. Epub 2003 Jun 13. Mol Biol Cell. 2003. PMID: 12972567 Free PMC article.
-
Maternal Ribosomes Are Sufficient for Tissue Diversification during Embryonic Development in C. elegans.Dev Cell. 2019 Mar 25;48(6):811-826.e6. doi: 10.1016/j.devcel.2019.01.019. Epub 2019 Feb 21. Dev Cell. 2019. PMID: 30799226 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

