Temperature sensitive nop2 alleles defective in synthesis of 25S rRNA and large ribosomal subunits in Saccharomyces cerevisiae
- PMID: 11452018
- PMCID: PMC55797
- DOI: 10.1093/nar/29.14.2927
Temperature sensitive nop2 alleles defective in synthesis of 25S rRNA and large ribosomal subunits in Saccharomyces cerevisiae
Abstract
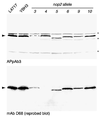
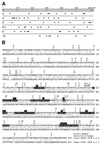
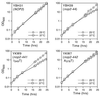
Using molecular genetic techniques, we have generated and characterized six temperature sensitive (ts) alleles of nop2. All failed to support growth at 37 degrees C and one was also formamide sensitive (fs) and failed to grow on media containing 3% formamide. Conditional lethality is not due to rapid turnover of mutant Nop2p proteins at 37 degrees C. Each allele contains between seven and 14 amino acid substitutions and one possesses a nonsense mutation near the C-terminus. Mapping experiments with one allele, nop2-4, revealed that a subset of the amino acid substitutions conferred the ts phenotype and that these mutations have an additive effect. All six mutants exhibited dramatic reductions in levels of 60S ribosome subunits under non-permissive conditions as well as some reduction at permissive temperature. Processing of 27S pre-rRNA to mature 25S rRNA was defective in all six mutants grown under non-permissive conditions. Levels of the 40S ribosomal subunit and 18S rRNA were not significantly affected. Amino acid substitutions in nop2 conditional alleles are discussed in the context of the hypothesis that Nop2p functions both as an RNA methyltransferase and a trans-acting factor in rRNA processing and large ribosomal subunit biogenesis.
Figures





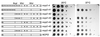
Similar articles
-
Nucleolar protein Nop12p participates in synthesis of 25S rRNA in Saccharomyces cerevisiae.Nucleic Acids Res. 2001 Jul 15;29(14):2938-49. doi: 10.1093/nar/29.14.2938. Nucleic Acids Res. 2001. PMID: 11452019 Free PMC article.
-
Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast.Mol Cell Biol. 1997 Jan;17(1):378-88. doi: 10.1128/MCB.17.1.378. Mol Cell Biol. 1997. PMID: 8972218 Free PMC article.
-
Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation.RNA. 2004 May;10(5):813-27. doi: 10.1261/rna.5255804. RNA. 2004. PMID: 15100437 Free PMC article.
-
Synthetic lethality with conditional dbp6 alleles identifies rsa1p, a nucleoplasmic protein involved in the assembly of 60S ribosomal subunits.Mol Cell Biol. 1999 Dec;19(12):8633-45. doi: 10.1128/MCB.19.12.8633. Mol Cell Biol. 1999. PMID: 10567587 Free PMC article.
-
RRP20, a component of the 90S preribosome, is required for pre-18S rRNA processing in Saccharomyces cerevisiae.Nucleic Acids Res. 2003 May 15;31(10):2524-33. doi: 10.1093/nar/gkg366. Nucleic Acids Res. 2003. PMID: 12736301 Free PMC article.
Cited by
-
Nop53p is a novel nucleolar 60S ribosomal subunit biogenesis protein.Biochem J. 2005 Jun 15;388(Pt 3):819-26. doi: 10.1042/BJ20041297. Biochem J. 2005. PMID: 15686447 Free PMC article.
-
5-methylcytosine in RNA: detection, enzymatic formation and biological functions.Nucleic Acids Res. 2010 Mar;38(5):1415-30. doi: 10.1093/nar/gkp1117. Epub 2009 Dec 8. Nucleic Acids Res. 2010. PMID: 20007150 Free PMC article. Review.
-
Nucleolar protein Nop12p participates in synthesis of 25S rRNA in Saccharomyces cerevisiae.Nucleic Acids Res. 2001 Jul 15;29(14):2938-49. doi: 10.1093/nar/29.14.2938. Nucleic Acids Res. 2001. PMID: 11452019 Free PMC article.
-
Rational extension of the ribosome biogenesis pathway using network-guided genetics.PLoS Biol. 2009 Oct;7(10):e1000213. doi: 10.1371/journal.pbio.1000213. Epub 2009 Oct 6. PLoS Biol. 2009. PMID: 19806183 Free PMC article.
-
The assembly factor Erb1 functions in multiple remodeling events during 60S ribosomal subunit assembly in S. cerevisiae.Nucleic Acids Res. 2017 May 5;45(8):4853-4865. doi: 10.1093/nar/gkw1361. Nucleic Acids Res. 2017. PMID: 28115637 Free PMC article.
References
-
- Ofengand J. and Fournier,M.J. (1998) The pseudouridine residues of rRNA: number, location, biosynthesis and function. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 229–253.
-
- Lowe T.M. and Eddy,S.R. (1999) A computational screen for methylation guide snoRNAs in yeast. Science, 283, 1168–1171. - PubMed
-
- Maden B.E.H. (1998) Intracellular locations of RNA-modifying enzymes. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 421–440.
-
- Tollervey D. and Kiss,T. (1997) Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol., 9, 337–342. - PubMed
-
- Grosjean H., Motorin,Y. and Morin,A. (1998) RNA-modifying and RNA-editing enzymes: methods for their identification. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 21–46.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

