Cloning and characterization of two guide RNA-binding proteins from mitochondria of Crithidia fasciculata: gBP27, a novel protein, and gBP29, the orthologue of Trypanosoma brucei gBP21
- PMID: 11452020
- PMCID: PMC55805
- DOI: 10.1093/nar/29.14.2950
Cloning and characterization of two guide RNA-binding proteins from mitochondria of Crithidia fasciculata: gBP27, a novel protein, and gBP29, the orthologue of Trypanosoma brucei gBP21
Abstract
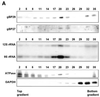
In kinetoplastid protozoa, mitochondrial (mt) mRNAs are post-transcriptionally edited by insertion and deletion of uridylate residues, the information being provided by guide (g)RNAs. Currently popular mechanisms for the editing process envisage a series of consecutive 'cut-and-paste' reactions, carried out by a complex RNP machinery. Here we report on the purification, cloning and functional analysis of two gRNA-binding proteins of 28.8 (gBP29) and 26.8 kDa (gBP27) from mitochondria of the insect trypanosome Crithidia fasciculata. gBP29 and gBP27 proved to be similar, Arg + Ala-rich proteins, with pI values of approximately 10.0. gBP27 has no homology to known proteins, but gBP29 is the C.fasciculata orthologue of gBP21 from Trypanosoma brucei, a gRNA-binding protein that associates with active RNA editing complexes. As measured in UV cross-linking assays, His-tagged recombinant gBP29 and gBP27 bind to radiolabelled poly(U) and synthetic gRNAs, while competition experiments suggest a role for the gRNA 3'-(U)-tail in binding to these proteins. Immunoprecipitates of mt extracts generated with antibodies against gBP29 also contained gBP27 and vice versa. The immunoprecipitates further harbored a large proportion of the cellular content of four different gRNAs and of edited and pre-edited NADH dehydrogenase subunit 7 mRNAs, but only small amounts of mt rRNAs. In addition, the bulk of gBP29 and gBP27 co-eluted with gRNAs from gel filtration columns in the high molecular weight range. Together, these results suggest that the proteins are part of a large macromolecular complex(es). We infer that gBP29 and gBP27 are components of the C.fasciculata editing machinery that may interact with gRNAs.
Figures







Similar articles
-
Identification by UV cross-linking of oligo(U)-binding proteins in mitochondria of the insect trypanosomatid Crithidia fasciculata.Eur J Biochem. 1995 Feb 1;227(3):780-6. doi: 10.1111/j.1432-1033.1995.tb20201.x. Eur J Biochem. 1995. PMID: 7867638
-
Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity.J Biol Chem. 1997 Feb 7;272(6):3749-57. doi: 10.1074/jbc.272.6.3749. J Biol Chem. 1997. PMID: 9013632
-
A possible role for the guide RNA U-tail as a specificity determinant in formation of guide RNA-messenger RNA chimeras in mitochondrial extracts of Crithidia fasciculata.Mol Biochem Parasitol. 1995 Jul;73(1-2):211-22. doi: 10.1016/0166-6851(95)00119-l. Mol Biochem Parasitol. 1995. PMID: 8577329
-
RNA editing in trypanosome mitochondria: guidelines for models.FEBS Lett. 1993 Jun 28;325(1-2):146-51. doi: 10.1016/0014-5793(93)81431-x. FEBS Lett. 1993. PMID: 7685714 Review.
-
Insertional and deletional RNA editing in trypanosome mitochondria.Nucleic Acids Symp Ser. 1997;(36):15-8. Nucleic Acids Symp Ser. 1997. PMID: 9478193 Review.
Cited by
-
Mitochondrial RNA editing in trypanosomes: small RNAs in control.Biochimie. 2014 May;100:125-31. doi: 10.1016/j.biochi.2014.01.003. Epub 2014 Jan 17. Biochimie. 2014. PMID: 24440637 Free PMC article. Review.
-
Mitochondrial membrane complex that contains proteins necessary for tRNA import in Trypanosoma brucei.J Biol Chem. 2012 Mar 16;287(12):8892-903. doi: 10.1074/jbc.M111.300186. Epub 2012 Jan 20. J Biol Chem. 2012. PMID: 22267727 Free PMC article.
-
Trypanosoma brucei 20 S editosomes have one RNA substrate-binding site and execute RNA unwinding activity.J Biol Chem. 2012 Jul 27;287(31):26268-77. doi: 10.1074/jbc.M112.365916. Epub 2012 Jun 1. J Biol Chem. 2012. PMID: 22661715 Free PMC article.
-
Uridine insertion/deletion editing in trypanosomes: a playground for RNA-guided information transfer.Wiley Interdiscip Rev RNA. 2011 Sep-Oct;2(5):669-85. doi: 10.1002/wrna.82. Epub 2011 Mar 23. Wiley Interdiscip Rev RNA. 2011. PMID: 21823228 Free PMC article. Review.
-
Identification of novel components of Trypanosoma brucei editosomes.RNA. 2003 Apr;9(4):484-92. doi: 10.1261/rna.2194603. RNA. 2003. PMID: 12649499 Free PMC article.
References
-
- Arts G.J. and Benne,R. (1996) Mechanism and evolution of RNA editing in kinetoplastida. Biochim. Biophys. Acta, 1307, 39–54. - PubMed
-
- Hajduk S.L. and Sabatini,R.S. (1998) Mitochondrial mRNA editing in kinetoplastid mitochondria. In Grosjean,H. and Benne,R. (eds) Modification and Editing of RNA. ASM press, Washington, DC, pp. 377–393.
-
- Seiwert S.D., Heidmann,S. and Stuart,K. (1996) Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell, 84, 831–841. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

