Functionally antagonistic sequences are required for normal autoregulation of Drosophila tra-2 pre-mRNA splicing
- PMID: 11452026
- PMCID: PMC55796
- DOI: 10.1093/nar/29.14.3012
Functionally antagonistic sequences are required for normal autoregulation of Drosophila tra-2 pre-mRNA splicing
Abstract
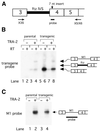
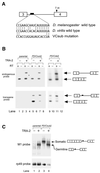
Expression of functional TRA-2 protein in the male germline of Drosophila is regulated through a negative feedback mechanism in which a specific TRA-2 isoform represses splicing of the M1 intron in the TRA-2 pre-mRNA. We have previously shown that the mechanism of M1 splicing repression is conserved between distantly related Drosophila species. Using transgenic fly strains, we have examined the effects on regulation of mutations in two conserved features of the M1 intron. Our results show that TRA-2-dependent repression of M1 splicing depends on the presence of a suboptimal non-consensus 3' splice site. Substitution of this 3' splice site with a strong splice site resulted in TRA-2 independent splicing, while substitution with an unrelated weak 3' splice site was compatible with repression, implying that reduced basal splicing efficiency is important for regulation. A second conserved element internal to the intron was found to be essential for efficient M1 splicing in the soma where the intron is not normally retained. We show that the role of this element is to enhance splicing and overcome the reduction in efficiency caused by the intron's suboptimal 3' splice site. Our results indicate that antagonistic elements in the M1 intron act together to establish a context that is permissive for repression of splicing by TRA-2 while allowing efficient splicing in the absence of a repressor.
Figures




Similar articles
-
Half pint/Puf68 is required for negative regulation of splicing by the SR splicing factor Transformer2.RNA Biol. 2013 Aug;10(8):1396-406. doi: 10.4161/rna.25645. Epub 2013 Jul 9. RNA Biol. 2013. PMID: 23880637 Free PMC article.
-
Autoregulation of transformer-2 alternative splicing is necessary for normal male fertility in Drosophila.Genetics. 1998 Jul;149(3):1477-86. doi: 10.1093/genetics/149.3.1477. Genetics. 1998. PMID: 9649535 Free PMC article.
-
Alternative pre-mRNA splicing in Drosophila spliceosomal assembly factor RNP-4F during development.Gene. 2006 Apr 26;371(2):234-45. doi: 10.1016/j.gene.2005.12.025. Epub 2006 Feb 23. Gene. 2006. PMID: 16497447
-
Molecular genetic aspects of sex determination in Drosophila melanogaster.Genome. 1989;31(2):638-45. doi: 10.1139/g89-117. Genome. 1989. PMID: 2517260 Review.
-
Pre-mRNA missplicing as a cause of human disease.Prog Mol Subcell Biol. 2006;44:27-46. doi: 10.1007/978-3-540-34449-0_2. Prog Mol Subcell Biol. 2006. PMID: 17076263 Review.
Cited by
-
The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior.Genes Dev. 2002 Nov 15;16(22):2879-92. doi: 10.1101/gad.1010302. Genes Dev. 2002. PMID: 12435630 Free PMC article.
-
Half pint/Puf68 is required for negative regulation of splicing by the SR splicing factor Transformer2.RNA Biol. 2013 Aug;10(8):1396-406. doi: 10.4161/rna.25645. Epub 2013 Jul 9. RNA Biol. 2013. PMID: 23880637 Free PMC article.
-
The doublesex splicing enhancer components Tra2 and Rbp1 also repress splicing through an intronic silencer.Mol Cell Biol. 2007 Jan;27(2):699-708. doi: 10.1128/MCB.01572-06. Epub 2006 Nov 13. Mol Cell Biol. 2007. PMID: 17101798 Free PMC article.
-
The Wright stuff: reimagining path analysis reveals novel components of the sex determination hierarchy in Drosophila melanogaster.BMC Syst Biol. 2015 Sep 4;9:53. doi: 10.1186/s12918-015-0200-0. BMC Syst Biol. 2015. PMID: 26335107 Free PMC article.
-
Concentration dependent selection of targets by an SR splicing regulator results in tissue-specific RNA processing.Nucleic Acids Res. 2006;34(21):6256-63. doi: 10.1093/nar/gkl755. Epub 2006 Nov 10. Nucleic Acids Res. 2006. PMID: 17098939 Free PMC article.
References
-
- Chabot B. (1996) Directing alternative splicing: cast and scenarios. Trends Genet., 12, 472–478. - PubMed
-
- Mattox W. and Baker,B.S. (1991) Autoregulation of the splicing of transcripts from the transformer-2 gene of Drosophila. Genes Dev., 5, 786–796. - PubMed
-
- Bell L.R., Horabin,J.I., Schedl,P. and Cline,T.W. (1991) Positive autoregulation of sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell, 65, 229–239. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

