Tripartite structure of Saccharomyces cerevisiae Dna2 helicase/endonuclease
- PMID: 11452032
- PMCID: PMC55803
- DOI: 10.1093/nar/29.14.3069
Tripartite structure of Saccharomyces cerevisiae Dna2 helicase/endonuclease
Abstract
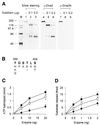
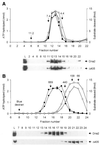
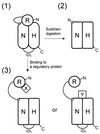
In order to gain insights into the structural basis of the multifunctional Dna2 enzyme involved in Okazaki fragment processing, we performed biochemical, biophysical and genetic studies to dissect the domain structure of Dna2. Proteolytic digestion of Dna2 using subtilisin produced a 127 kDa polypeptide that lacked the 45 kDa N-terminal region of Dna2. Further digestion generated two subtilisin-resistant core fragments of approximately equal size, 58 and 60 kDa. Surprisingly, digestion resulted in a significant (3- to 8-fold) increase in both ATPase and endonuclease activities compared to the intact enzyme. However, cells with a mutant DNA2 allele lacking the corresponding N-terminal region were severely impaired in growth, being unable to grow at 37 degrees C, indicating that the N-terminal region contains a domain critical for a cellular function(s) of Dna2. Analyses of the hydrodynamic properties of and in vivo complex formation by wild-type and/or mutant Dna2 lacking the N-terminal 45 kDa domain revealed that Dna2 is active as the monomer and thus the defect in the mutant Dna2 protein is not due to its inability to multimerize. In addition, we found that the N-terminal 45 kDa domain interacts physically with a central region located between the two catalytic domains. Our results suggest that the N-terminal 45 kDa domain of Dna2 plays a critical role in regulation of the enzymatic activities of Dna2 by serving as a site for intra- and intermolecular interactions essential for optimal function of Dna2 in Okazaki fragment processing. The possible mode of regulation of Dna2 is discussed based upon our recent finding that replication protein A interacts functionally and physically with Dna2 during Okazaki fragment processing.
Figures
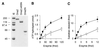





References
-
- Bae S.H. and Seo,Y.S. (2000) Characterization of the enzymatic properties of the yeast dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem., 275, 38022–38031. - PubMed
-
- Budd M.E. and Campbell,J.L. (2000) The pattern of sensitivity of yeast dna2 mutants to DNA damaging agents suggests a role in DSB and postreplication repair pathways. Mutat. Res., 459, 173–186. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

