Biochemical characterization of a novel hypoxanthine/xanthine dNTP pyrophosphatase from Methanococcus jannaschii
- PMID: 11452035
- PMCID: PMC55802
- DOI: 10.1093/nar/29.14.3099
Biochemical characterization of a novel hypoxanthine/xanthine dNTP pyrophosphatase from Methanococcus jannaschii
Abstract
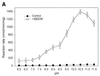
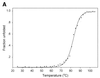
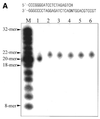
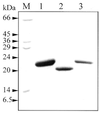
A novel dNTP pyrophosphatase, Mj0226 from Methanococcus jannaschii, which catalyzes the hydrolysis of nucleoside triphosphates to the monophosphate and PPi, has been characterized. Mj0226 protein catalyzes hydrolysis of two major substrates, dITP and XTP, suggesting that the 6-keto group of hypoxanthine and xanthine is critical for interaction with the protein. Under optimal reaction conditions the k(ca)(t) /K(m) value for these substrates was approximately 10 000 times that with dATP. Neither endonuclease nor 3'-exonuclease activities were detected in this protein. Interestingly, dITP was efficiently inserted opposite a dC residue in a DNA template and four dNTPs were also incorporated opposite a hypoxanthine residue in template DNA by DNA polymerase I. Two protein homologs of Mj0226 from Escherichia coli and Archaeoglobus fulgidus were also cloned and purified. These have catalytic activities similar to Mj0226 protein under optimal conditions. The implications of these results have significance in understanding how homologous proteins, including Mj0226, act biologically in many organisms. It seems likely that Mj0226 and its homologs have a major role in preventing mutations caused by incorporation of dITP and XTP formed spontaneously in the nucleotide pool into DNA. This report is the first identification and functional characterization of an enzyme hydrolyzing non-canonical nucleotides, dITP and XTP.
Figures






References
-
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
-
- Shapiro R. and Pohl,S.H. (1968) The reaction of ribonucleosides with nitrous acid. Side products and kinetics. Biochemistry, 7, 448–455. - PubMed
-
- Lindahl T. (1979) DNA glycosylases, endonucleases for apurinic/apyrimidinic sites and base excision-repair. Prog. Nucleic Acid Res. Mol. Biol., 22, 135–192. - PubMed
-
- Schuster H. (1960) The reaction of nitrous acid with deoxyribonucleic acid. Biochem. Biophys. Res. Commun., 2, 320–323.
-
- Hill-Perkins M., Jones,M.D. and Karran,P. (1986) Site-specific mutagenesis in vivo by single methylated or deaminated purine bases. Mutat. Res., 162, 153–163. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

