A homogeneous europium cryptate-based assay for the diagnosis of mutations by time-resolved fluorescence resonance energy transfer
- PMID: 11452039
- PMCID: PMC55817
- DOI: 10.1093/nar/29.14.e70
A homogeneous europium cryptate-based assay for the diagnosis of mutations by time-resolved fluorescence resonance energy transfer
Abstract
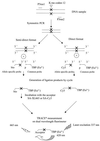
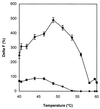
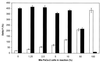
Oligonucleotide ligation assay (OLA) is considered to be a very useful methodology for the detection and characterization of mutations, particularly for clinical purposes. The fluorescence resonance energy transfer between a fluorescent donor and a suitable fluorophore as acceptor has been applied in the past to several scientific fields. This technique is well adapted to nucleic acid analysis such as DNA sequencing, DNA hybridization and polymerase chain reaction. We describe here a homogeneous format based on the use of a rare earth cryptate label as donor: tris-bipyridine-Eu(3+). The long-lived fluorescence of this label makes it possible to reach a high sensitivity by using a time-resolved detection mode. A non-radiative energy transfer technology, known as time-resolved amplification of cryptate emission (TRACE((R))) characterized by a temporal and spectral selectivity has been developed. The TRACE((R)) detection of characterized single nucleotide polymorphism using the OLA for allelic discrimination is proposed. We demonstrate the potentialities of this OLA-TRACE((R)) methodology through the analysis of K-ras oncogene point mutations.
Figures
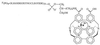

Similar articles
-
Europium cryptate-tethered ribonucleotide for the labeling of RNA and its detection by time-resolved amplification of cryptate emission.Anal Biochem. 2000 Nov 1;286(1):17-25. doi: 10.1006/abio.2000.4764. Anal Biochem. 2000. PMID: 11038268
-
A homogeneous time-resolved fluorescence detection of telomerase activity.Anal Biochem. 2004 Oct 1;333(1):105-13. doi: 10.1016/j.ab.2004.06.006. Anal Biochem. 2004. PMID: 15351286
-
Europium cryptate labeled deoxyuridine-triphosphate analog: synthesis and enzymatic incorporation.Nucleosides Nucleotides Nucleic Acids. 2000 Sep;19(9):1463-74. doi: 10.1080/15257770008033854. Nucleosides Nucleotides Nucleic Acids. 2000. PMID: 11092315
-
Time resolved amplification of cryptate emission: a versatile technology to trace biomolecular interactions.J Biotechnol. 2002 Jan;82(3):233-50. doi: 10.1016/s1389-0352(01)00040-x. J Biotechnol. 2002. PMID: 11999692 Review.
-
Homogeneous time resolved fluorescence resonance energy transfer using rare earth cryptates as a tool for probing molecular interactions in biology.Spectrochim Acta A Mol Biomol Spectrosc. 2001 Sep 14;57(11):2197-211. doi: 10.1016/s1386-1425(01)00493-0. Spectrochim Acta A Mol Biomol Spectrosc. 2001. PMID: 11603838 Review.
Cited by
-
Luminescent Lanthanides in Biorelated Applications: From Molecules to Nanoparticles and Diagnostic Probes to Therapeutics.Chem Rev. 2025 Feb 26;125(4):2269-2370. doi: 10.1021/acs.chemrev.4c00615. Epub 2025 Feb 17. Chem Rev. 2025. PMID: 39960048 Free PMC article. Review.
-
Rapid genotyping using pyrene-perylene locked nucleic acid complexes.Artif DNA PNA XNA. 2013 Apr-Jun;4(2):58-68. doi: 10.4161/adna.25903. Artif DNA PNA XNA. 2013. PMID: 24044052 Free PMC article.
-
A Time-Resolved FRET Assay Identifies a Small Molecule that Inhibits the Essential Bacterial Cell Wall Polymerase FtsW.Angew Chem Int Ed Engl. 2023 Jun 19;62(25):e202301522. doi: 10.1002/anie.202301522. Epub 2023 May 11. Angew Chem Int Ed Engl. 2023. PMID: 37099323 Free PMC article.
-
A time-resolved Förster resonance energy transfer assay to measure activity of the deamidase of the prokaryotic ubiquitin-like protein.Anal Biochem. 2015 Oct 15;487:27-9. doi: 10.1016/j.ab.2015.07.003. Epub 2015 Jul 21. Anal Biochem. 2015. PMID: 26205584 Free PMC article.
-
Review: immunoassays in DNA damage and instability detection.Cell Mol Life Sci. 2019 Dec;76(23):4689-4704. doi: 10.1007/s00018-019-03239-6. Epub 2019 Jul 24. Cell Mol Life Sci. 2019. PMID: 31342119 Free PMC article. Review.
References
-
- Saiki R.K., Scharf,S., Faloona,F., Mullis,K.B., Horn,G.T., Erlich,H.A. and Arnheim,N. (1985) Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science, 230, 1350–1354. - PubMed
-
- Myers R.M., Larin,Z. and Maniatis,T. (1985) Detection of single base substitutions by ribonuclease cleavage at mismatches in RNA:DNA duplexes. Science, 230, 1242–1246. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

