The solution structure of PapGII from uropathogenic Escherichia coli and its recognition of glycolipid receptors
- PMID: 11454740
- PMCID: PMC1083947
- DOI: 10.1093/embo-reports/kve133
The solution structure of PapGII from uropathogenic Escherichia coli and its recognition of glycolipid receptors
Abstract
Uropathogenic Escherichia coli (UPEC) is the primary cause of symptomatic urinary tract infection. The P-pili, a bacterial surface organelle, mediates the bacterial host--cell adhesion. The PapG adhesin has generated much interest in recent years, not only because of its clinical value, i.e. in the prevention of microbial adherence, but also because of its ability to promote virulence. Using multidimensional nuclear magnetic resonance (NMR) and deuteration we have determined the solution structure of the adhesin domain from PapGII (PapGII-198). The novel structure of PapGII-198 is composed of a large elongated jellyroll motif. Despite an automated search of the structural database failing to reveal any similar proteins, PapGII adhesin shares some structural similarities with FimH. Furthermore, interpretation of NMR-titration data has enabled us to identify the putative binding site for the globoseries of oligosaccharides. This work provides insight into UPEC pathogenesis as well as aiding the development of preventative therapies and the guidance of future mutagenesis programmes.
Figures
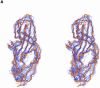
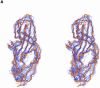
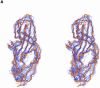
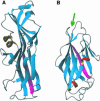

Similar articles
-
Quantitative studies of the binding of the class II PapG adhesin from uropathogenic Escherichia coli to oligosaccharides.Bioorg Med Chem. 2003 May 15;11(10):2255-61. doi: 10.1016/s0968-0896(03)00114-7. Bioorg Med Chem. 2003. PMID: 12713835
-
Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor.Cell. 2001 Jun 15;105(6):733-43. doi: 10.1016/s0092-8674(01)00388-9. Cell. 2001. PMID: 11440716
-
The fimbrial adhesin F17-G of enterotoxigenic Escherichia coli has an immunoglobulin-like lectin domain that binds N-acetylglucosamine.Mol Microbiol. 2003 Aug;49(3):705-15. doi: 10.1046/j.1365-2958.2003.03600.x. Mol Microbiol. 2003. PMID: 12864853
-
Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney.Kidney Int. 2007 Jul;72(1):19-25. doi: 10.1038/sj.ki.5002230. Epub 2007 Mar 28. Kidney Int. 2007. PMID: 17396114 Review.
-
Molecular structure of adhesin domains in Escherichia coli fimbriae.Int J Med Microbiol. 2005 Oct;295(6-7):479-86. doi: 10.1016/j.ijmm.2005.06.010. Int J Med Microbiol. 2005. PMID: 16238022 Review.
Cited by
-
Mapping the binding domain of the F18 fimbrial adhesin.Infect Immun. 2003 Apr;71(4):2163-72. doi: 10.1128/IAI.71.4.2163-2182.2003. Infect Immun. 2003. PMID: 12654838 Free PMC article.
-
Dimeric and Trimeric Fusion Proteins Generated with Fimbrial Adhesins of Uropathogenic Escherichia coli.Front Cell Infect Microbiol. 2016 Oct 31;6:135. doi: 10.3389/fcimb.2016.00135. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27843814 Free PMC article.
-
Structural Diversities of Lectins Binding to the Glycosphingolipid Gb3.Front Mol Biosci. 2021 Jul 26;8:704685. doi: 10.3389/fmolb.2021.704685. eCollection 2021. Front Mol Biosci. 2021. PMID: 34381814 Free PMC article. Review.
-
Differential carbohydrate recognition by Campylobacter jejuni strain 11168: influences of temperature and growth conditions.PLoS One. 2009;4(3):e4927. doi: 10.1371/journal.pone.0004927. Epub 2009 Mar 17. PLoS One. 2009. PMID: 19290056 Free PMC article.
-
Alternative Therapeutic Options to Antibiotics for the Treatment of Urinary Tract Infections.Front Microbiol. 2020 Jul 3;11:1509. doi: 10.3389/fmicb.2020.01509. eCollection 2020. Front Microbiol. 2020. PMID: 32719668 Free PMC article. Review.
References
-
- Bax A., Clore, G.M. and Gronenborn, A.M. (1990) H-1-H-1 correlation via isotropic mixing of C-13 magnetization, a new 3-dimensional approach for assigning H-1 and C-13 spectra of C-13-enriched proteins. J. Magn. Reson., 88, 425–431.
-
- Bizebard T., Gigant, B., Rigolet, P., Rasmussen, B., Diat, O., Bosecke, P., Wharton, S.A., Skehel, J.J. and Knossow, M. (1995) Structure of influenza-virus hemagglutinin complexed with a neutralizing antibody. Nature, 376, 92–94. - PubMed
-
- Bock K. et al. (1985) Specificity of binding of a strain of uropathogenic Escherichia coli to Galα1–4Gal-containing glycosphingolipids. J. Biol. Chem., 260, 8545–8551. - PubMed
-
- Brunger A.T. et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
-
- Choudhury D., Thompson, A., Stojanoff, V., Langermann, S., Pinkner, J., Hultgren, S.J. and Knight, S.D. (1999) X-ray structure of the FimC–FimH chaperone–adhesin complex from uropathogenic Escherichia coli. Science, 285, 1061–1066. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

