Color transitions in coral's fluorescent proteins by site-directed mutagenesis
- PMID: 11459517
- PMCID: PMC34604
- DOI: 10.1186/1471-2091-2-6
Color transitions in coral's fluorescent proteins by site-directed mutagenesis
Abstract
Background: Green Fluorescent Protein (GFP) cloned from jellyfish Aequorea victoria and its homologs from corals Anthozoa have a great practical significance as in vivo markers of gene expression. Also, they are an interesting puzzle of protein science due to an unusual mechanism of chromophore formation and diversity of fluorescent colors. Fluorescent proteins can be subdivided into cyan (approximately 485 nm), green (approximately 505 nm), yellow (approximately 540 nm), and red (>580 nm) emitters.
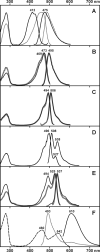
Results: Here we applied site-directed mutagenesis in order to investigate the structural background of color variety and possibility of shifting between different types of fluorescence. First, a blue-shifted mutant of cyan amFP486 was generated. Second, it was established that cyan and green emitters can be modified so as to produce an intermediate spectrum of fluorescence. Third, the relationship between green and yellow fluorescence was inspected on closely homologous green zFP506 and yellow zFP538 proteins. The following transitions of colors were performed: yellow to green; yellow to dual color (green and yellow); and green to yellow. Fourth, we generated a mutant of cyan emitter dsFP483 that demonstrated dual color (cyan and red) fluorescence.
Conclusions: Several amino acid substitutions were found to strongly affect fluorescence maxima. Some positions primarily found by sequence comparison were proved to be crucial for fluorescence of particular color. These results are the first step towards predicting the color of natural GFP-like proteins corresponding to newly identified cDNAs from corals.
Figures

References
-
- Tsien RY, Prasher D. Molecular biology and mutation of green fluorescent protein. Green fluorescent protein. Chalfie, M., Kain, S., Eds. Willey-Liss, New York, 1998:97–118.
-
- Palm GJ, Zdanov A, Gaitanaris GA, Stauber R, Pavlakis GN, Wlodawer A. The structural basis for spectral variations in green fluorescent protein. Nature Struct Biol. 1997;4:361–365. - PubMed
-
- Remington SJ. Structural basis for understanding spectral variations in green fluorescent protein. Methods Enzymol. 2000;305:196–211. - PubMed
-
- Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Current Biol. 1996;6:178–182. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

