Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene
- PMID: 11459824
- PMCID: PMC312734
- DOI: 10.1101/gad.905701
Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene
Abstract
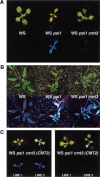
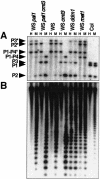
Plants maintain cytosine methylation at CG and non-CG residues to control gene expression and genome stability. In a screen for Arabidopsis mutants that alter methylation and silencing of a densely methylated endogenous reporter gene, we recovered 11 loss-of-function alleles in the CMT3 chromomethylase gene. The cmt3 mutants displayed enhanced expression and reduced methylation of the reporter, particularly at non-CG cytosines. CNG methylation was also reduced at repetitive centromeric sequences. Thus, CMT3 is a key determinant for non-CG methylation. The lack of CMT homologs in animal genomes could account for the observation that in contrast to plants, animals maintain primarily CG methylation.
Figures


References
-
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. - PubMed
-
- Bender J, Fink GR. Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell. 1995;83:725–734. - PubMed
-
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
