Capping, splicing, and 3' processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD
- PMID: 11459828
- PMCID: PMC312735
- DOI: 10.1101/gad.889101
Capping, splicing, and 3' processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD
Abstract
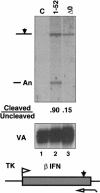
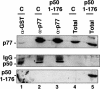
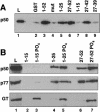
Capping, splicing, and cleavage/polyadenylation of pre-mRNAs are interdependent events that are all stimulated in vivo by the carboxy-terminal domain (CTD) of RNA Pol II. We show that the CTD independently enhances splicing and 3' processing and that stimulation of splicing by enhancers is facilitated by the CTD. We provide evidence that stimulation of 3' processing by the CTD requires contact with the 50-kD subunit of the cleavage stimulation factor, CstF. Overexpression of the CTD-binding domain of CstF p50 had a dominant-negative effect on 3' processing without disrupting the CstF complex. The CTD comprises 52 heptad repeats. The CTD carboxyl terminus including heptads 27-52 supported capping, splicing, and 3' processing but the amino terminus supported only capping. We conclude that the CTD independently stimulates all three major pre-mRNA processing steps and that different regions of the CTD can serve distinct functions in pre-mRNA processing.
Figures



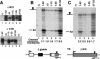
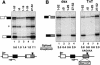


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
