Single-strand interruptions in replicating chromosomes cause double-strand breaks
- PMID: 11459959
- PMCID: PMC37427
- DOI: 10.1073/pnas.131009198
Single-strand interruptions in replicating chromosomes cause double-strand breaks
Abstract
Replication-dependent chromosomal breakage suggests that replication forks occasionally run into nicks in template DNA and collapse, generating double-strand ends. To model replication fork collapse in vivo, I constructed phage lambda chromosomes carrying the nicking site of M13 bacteriophage and infected with these substrates Escherichia coli cells, producing M13 nicking enzyme. I detected double-strand breaks at the nicking sites in lambda DNA purified from these cells. The double-strand breakage depends on (i) the presence of the nicking site; (ii) the production of the nicking enzyme; and (iii) replication of the nick-containing chromosome. Replication fork collapse at nicks in template DNA explains diverse phenomena, including eukaryotic cell killing by DNA topoisomerase inhibitors and inviability of recombination-deficient vertebrate cell lines.
Figures
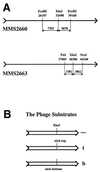
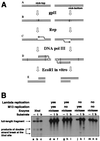
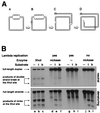

References
-
- Ginsberg D M, Jagger J. J Gen Microbiol. 1965;40:171–184. - PubMed
-
- Hanawalt P C. Photochem Photobiol. 1966;5:1–12. - PubMed
-
- Li L H, Fraser T J, Olin E J, Bhuyan B K. Cancer Res. 1972;32:2643–2650. - PubMed
-
- Tyrrell R M, Moss S H, Davies D J G. Mutat Res. 1972;16:1–12. - PubMed
-
- Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

