Bacteriophage T4 gene 41 helicase and gene 59 helicase-loading protein: a versatile couple with roles in replication and recombination
- PMID: 11459969
- PMCID: PMC37437
- DOI: 10.1073/pnas.121009398
Bacteriophage T4 gene 41 helicase and gene 59 helicase-loading protein: a versatile couple with roles in replication and recombination
Abstract
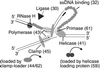
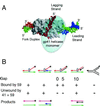
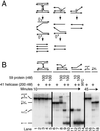
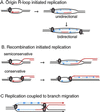
Bacteriophage T4 uses two modes of replication initiation: origin-dependent replication early in infection and recombination-dependent replication at later times. The same relatively simple complex of T4 replication proteins is responsible for both modes of DNA synthesis. Thus the mechanism for loading the T4 41 helicase must be versatile enough to allow it to be loaded on R loops created by transcription at several origins, on D loops created by recombination, and on stalled replication forks. T4 59 helicase-loading protein is a small, basic, almost completely alpha-helical protein whose N-terminal domain has structural similarity to high mobility group family proteins. In this paper we review recent evidence that 59 protein recognizes specific structures rather than specific sequences. It binds and loads the helicase on replication forks and on three- and four-stranded (Holliday junction) recombination structures, without sequence specificity. We summarize our experiments showing that purified T4 enzymes catalyze complete unidirectional replication of a plasmid containing the T4 ori(uvsY) origin, with a preformed R loop at the position of the R loop identified at this origin in vivo. This replication depends on the 41 helicase and is strongly stimulated by 59 protein. Moreover, the helicase-loading protein helps to coordinate leading and lagging strand synthesis by blocking replication on the ori(uvsY) R loop plasmid until the helicase is loaded. The T4 enzymes also can replicate plasmids with R loops that do not have a T4 origin sequence, but only if the R loops are within an easily unwound DNA sequence.
Figures



References
-
- Kreuzer K N, Morrical S W. In: Molecular Biology of Bacteriophage T4. Karem J, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 28–42.
-
- Kreuzer K N. Trends Biochem Sci. 2000;25:165–173. - PubMed
-
- Mosig G, Colowick N, Gruidl M E, Chang A, Harvey A J. FEMS Microbiol Rev. 1995;17:83–98. - PubMed
-
- Mosig G. Annu Rev Genet. 1998;32:379–413. - PubMed
-
- Nossal N G. In: Molecular Biology of Bacteriophage T4. Karem J, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 43–53.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

