A human rotavirus with rearranged genes 7 and 11 encodes a modified NSP3 protein and suggests an additional mechanism for gene rearrangement
- PMID: 11462002
- PMCID: PMC114965
- DOI: 10.1128/JVI.75.16.7305-7314.2001
A human rotavirus with rearranged genes 7 and 11 encodes a modified NSP3 protein and suggests an additional mechanism for gene rearrangement
Abstract
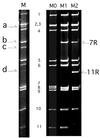
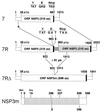
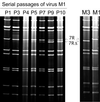
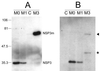
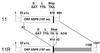
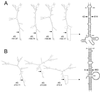
A human rotavirus (isolate M) with an atypical electropherotype with 14 apparent bands of double-stranded RNA was isolated from a chronically infected immunodeficient child. MA-104 cell culture adaptation showed that the M isolate was a mixture of viruses containing standard genes (M0) or rearranged genes: M1 (containing a rearranged gene 7) and M2 (containing rearranged genes 7 and 11). The rearranged gene 7 of virus M1 (gene 7R) was very unusual because it contained two complete open reading frames (ORF). Moreover, serial propagation of virus M1 in cell culture indicated that gene 7R rapidly evolved, leading to a virus with a deleted gene 7R (gene 7RDelta). Gene 7RDelta coded for a modified NSP3 protein (NSP3m) of 599 amino acids (aa) containing a repetition of aa 8 to 296. The virus M3 (containing gene 7RDelta) was not defective in cell culture and actually produced NSP3m. The rearranged gene 11 (gene 11R) had a more usual pattern, with a partial duplication leading to a normal ORF followed by a long 3' untranslated region. The rearrangement in gene 11R was almost identical to some of those previously described, suggesting that there is a hot spot for gene rearrangements at a specific location on the sequence. It has been suggested that in some cases the existence of short direct repeats could favor the occurrence of rearrangement at a specific site. The computer modeling of gene 7 and 11 mRNAs led us to propose a new mechanism for gene rearrangements in which secondary structures, besides short direct repeats, might facilitate and direct the transfer of the RNA polymerase from the 5' to the 3' end of the plus-strand RNA template during the replication step.
Figures
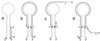
References
-
- Allen A M, Desselberger U. Reassortment of human rotaviruses carrying rearranged genomes with bovine rotavirus. J Gen Virol. 1985;66:2703–2714. - PubMed
-
- Aponte C, Mattion N M, Estes M K, Charpilienne A, Cohen J. Expression of two bovine rotavirus non-structural proteins (NSP2, NSP3) in the baculovirus system and production of monoclonal antibodies directed against the expressed proteins. Arch Virol. 1993;133:85–95. - PubMed
-
- Ballard A, McCrae M A, Desselberger U. Nucleotide sequences of normal and rearranged RNA segments 10 of human rotaviruses. J Gen Virol. 1992;73:633–638. - PubMed
-
- Besselaar T G, Rosenblatt A, Kidd A H. Atypical rotavirus from South African neonates. Arch Virol. 1986;87:327–330. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

