Proteolysis of monomeric recombinant rotavirus VP4 yields an oligomeric VP5* core
- PMID: 11462006
- PMCID: PMC114969
- DOI: 10.1128/JVI.75.16.7339-7350.2001
Proteolysis of monomeric recombinant rotavirus VP4 yields an oligomeric VP5* core
Abstract
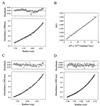
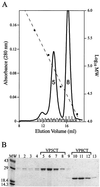
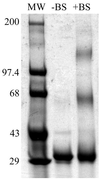
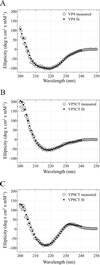
Rotavirus particles are activated for cell entry by trypsin cleavage of the outer capsid spike protein, VP4, into a hemagglutinin, VP8*, and a membrane penetration protein, VP5*. We have purified rhesus rotavirus VP4, expressed in baculovirus-infected insect cells. Purified VP4 is a soluble, elongated monomer, as determined by analytical ultracentrifugation. Trypsin cleaves purified VP4 at a number of sites that are protected on the virion and yields a heterogeneous group of protease-resistant cores of VP5*. The most abundant tryptic VP5* core is trimmed past the N terminus associated with activation for virus entry into cells. Sequential digestion of purified VP4 with chymotrypsin and trypsin generates homogeneous VP8* and VP5* cores (VP8CT and VP5CT, respectively), which have the authentic trypsin cleavages in the activation region. VP8CT is a soluble monomer composed primarily of beta-sheets. VP5CT forms sodium dodecyl sulfate-resistant dimers. These results suggest that trypsinization of rotavirus particles triggers a rearrangement in the VP5* region of VP4 to yield the dimeric spikes observed in icosahedral image reconstructions from electron cryomicroscopy of trypsinized rotavirus virions. The solubility of VP5CT and of trypsinized rotavirus particles suggests that the trypsin-triggered conformational change primes VP4 for a subsequent rearrangement that accomplishes membrane penetration. The domains of VP4 defined by protease analysis contain all mapped neutralizing epitopes, sialic acid binding residues, the heptad repeat region, and the membrane permeabilization region. This biochemical analysis of VP4 provides sequence-specific structural information that complements electron cryomicroscopy data and defines targets and strategies for atomic-resolution structural studies.
Figures


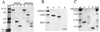
Similar articles
-
Localization of membrane permeabilization and receptor binding sites on the VP4 hemagglutinin of rotavirus: implications for cell entry.J Mol Biol. 2001 Dec 14;314(5):985-92. doi: 10.1006/jmbi.2000.5238. J Mol Biol. 2001. PMID: 11743716
-
A rotavirus spike protein conformational intermediate binds lipid bilayers.J Virol. 2010 Feb;84(4):1764-70. doi: 10.1128/JVI.01682-09. Epub 2009 Dec 9. J Virol. 2010. PMID: 20007281 Free PMC article.
-
Trypsin cleavage stabilizes the rotavirus VP4 spike.J Virol. 2001 Jul;75(13):6052-61. doi: 10.1128/JVI.75.13.6052-6061.2001. J Virol. 2001. PMID: 11390607 Free PMC article.
-
Rotavirus Replication: Gaps of Knowledge on Virus Entry and Morphogenesis.Tohoku J Exp Med. 2019 Aug;248(4):285-296. doi: 10.1620/tjem.248.285. Tohoku J Exp Med. 2019. PMID: 31447474 Review.
-
Proteolytic enhancement of human rotavirus infectivity.Clin Infect Dis. 1993 Mar;16 Suppl 2:S92-7. doi: 10.1093/clinids/16.supplement_2.s92. Clin Infect Dis. 1993. PMID: 8384014 Review.
Cited by
-
Specificity and affinity of sialic acid binding by the rhesus rotavirus VP8* core.J Virol. 2002 Oct;76(20):10512-7. doi: 10.1128/jvi.76.20.10512-10517.2002. J Virol. 2002. PMID: 12239329 Free PMC article.
-
Rhesus rotavirus entry into a polarized epithelium is endocytosis dependent and involves sequential VP4 conformational changes.J Virol. 2011 Mar;85(6):2492-503. doi: 10.1128/JVI.02082-10. Epub 2010 Dec 29. J Virol. 2011. PMID: 21191022 Free PMC article.
-
Using Species a Rotavirus Reverse Genetics to Engineer Chimeric Viruses Expressing SARS-CoV-2 Spike Epitopes.J Virol. 2022 Jul 27;96(14):e0048822. doi: 10.1128/jvi.00488-22. Epub 2022 Jun 27. J Virol. 2022. PMID: 35758692 Free PMC article.
-
Interactions of rotavirus VP4 spike protein with the endosomal protein Rab5 and the prenylated Rab acceptor PRA1.J Virol. 2003 Jun;77(12):7041-7. doi: 10.1128/jvi.77.12.7041-7047.2003. J Virol. 2003. PMID: 12768023 Free PMC article.
-
Rafts promote assembly and atypical targeting of a nonenveloped virus, rotavirus, in Caco-2 cells.J Virol. 2002 May;76(9):4591-602. doi: 10.1128/jvi.76.9.4591-4602.2002. J Virol. 2002. PMID: 11932424 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

