Accumulation and intranuclear distribution of unintegrated human immunodeficiency virus type 1 DNA
- PMID: 11462040
- PMCID: PMC115003
- DOI: 10.1128/JVI.75.16.7683-7691.2001
Accumulation and intranuclear distribution of unintegrated human immunodeficiency virus type 1 DNA
Abstract
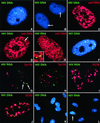
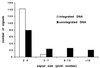
The RNA genome of human immunodeficiency virus type 1 (HIV-1) is converted into DNA after infection in order to integrate into the host cell DNA. However, a large number of these reverse-transcribed genomes remain unintegrated in the nucleus of infected cells. Currently, there are no data available about the intranuclear distribution pattern of unintegrated HIV-1 DNA in relation to nuclear structures as observed on the single-cell level. In the present study, we investigated the intranuclear fate of unintegrated viral DNA in cell lines expressing CD4 and coreceptors (HOS-CD4.CCR5 and U373-MAGI-CXCR4(CEM)) infected with HIV-1 (strain 89.6). We used a novel approach to distinguish in situ unintegrated from integrated viral DNA by performing fluorescent in situ hybridization on cells in which stress-induced chromosome condensation had been induced, a procedure that contracts chromosomes independent of the cell cycle. Cells infected for 15 h accumulated large amounts of HIV-1 DNA which was located between the condensed chromosome strands, allowing the identification of this viral DNA as unintegrated. In contrast, in HeLa/LAV, a cell line carrying integrated HIV-1 genomes, the great majority of viral DNA colocalized with the cellular DNA. We show that unintegrated HIV-1 DNA does not evenly distribute within the host cell nucleus but tends to aggregate into clusters containing many copies of the viral genomes. The formation of these DNA clusters was independent of viral DNA replication and thus appeared to result solely from multiple infections. The DNA aggregates remained in the nuclei of infected cells for at least 25 h after the infection was stopped. The emergence of transcription sites, which most likely denote sites of the integrated provirus, lagged clearly behind the accumulation of viral DNA. These transcription foci could not be linked to unintegrated DNA molecules, suggesting that this DNA type is unable to transcribe, at least at levels comparable to those of integrated DNA. Neither unintegrated HIV-1 DNA nor transcription foci nor integrated DNA was observed to associate with nuclear domain 10 (ND10), a nuclear structure known to represent the site where several DNA viruses replicate and transcribe. Also, HIV-1 does not modify ND10 at early or late times of infection. There was no specific association of HIV-1 transcripts with splicing factor SC35 domains, in contrast to what has been reported for a number of both cellular and viral genes. Surprisingly, unintegrated HIV-1 DNA was found to accumulate within or in close association with SC35 domains, demonstrating a specific distribution of the viral DNA within the host cell nucleus. Taken together, our results demonstrate that unintegrated proviral HIV-1 DNA does not randomly localize within infected cells but preferentially aggregates in the nucleus within SC35 domains.
Figures

References
-
- Barbosa P, Charneau P, Dumey N, Clavel F. Kinetic analysis of HIV-1 early replicative steps in a coculture system. AIDS Res Hum Retrovir. 1994;10:53–59. - PubMed
-
- Berg J, Doe B, Steimer K S, Wabl M. HeLa-LAV, an epithelial cell line stably infected with HIV-1. J Virol Methods. 1991;34:173–180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

