Demethylation, reactivation, and destabilization of human fragile X full-mutation alleles in mouse embryocarcinoma cells
- PMID: 11462172
- PMCID: PMC1235481
- DOI: 10.1086/322739
Demethylation, reactivation, and destabilization of human fragile X full-mutation alleles in mouse embryocarcinoma cells
Abstract
The major causes of fragile X syndrome are mutational expansion of the CGG repeat in the FMR1 gene, hypermethylation, and transcriptional silencing. Most fragile X embryos develop somatic mosaicism of disease-causing "full" expansions of different lengths. Homogeneity of the mosaic patterns among multiple tissues in the same individual indicates that these previously unstable expansions acquire mitotic stability early in fetal life. Since mitotic stability is found strictly associated with hypermethylation in adult tissues, current theory has fixed the time of instability to developmental stages when fully expanded CGG repeats exist in an unmethylated state. We used murine embryocarcinoma (EC) cells (PC13) as a model system of pluripotent embryonic cells. Hypermethylated and unmethylated full expansions on human fragile X chromosomes were transferred from murine A9 hybrids into EC cells, by means of microcell fusion. As demonstrated in the present study for the first time, even full expansion alleles that were fully methylated and stable in the donors' fibroblasts and in A9 became demethylated, reactivated, and destabilized in undifferentiated EC hybrids. When destabilized expansions were reintroduced from EC cells into A9, instability was reversed to stability. Our results strongly support the idea that fully expanded alleles are initially unstable and unmethylated in the human embryo and gain stability upon genetic or epigenetic change of the embryonic cells.
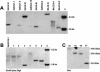
Figures




References
Electronic-Database Information
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human FMR1 mRNA sequence [accession number XM_010288])
-
- Jackson Laboratory, The, http://www.jax.org/ (for strain-specific marker alleles and PCR primers)
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for fragile X syndrome [MIM 309550])
References
-
- Bontekoe CJ, de Graaff E, Nieuwenhuizen IM, Willemsen R, Oostra BA (1997) FMR1 premutation allele (CGG)81 is stable in mice. Eur J Hum Genet 5:293–298 - PubMed
-
- Burman RW, Popovich BW, Jacky PB, Turker MS (1999) Fully expanded FMR1 CGG repeats exhibit a length- and differentiation-dependent instability in cell hybrids that is independent of DNA methylation. Hum Mol Genet 8:2293–2302 - PubMed
-
- Chiurazzi P, Pomponi MG, Pietrobono R, Bakker CE, Neri G, Oostra BA (1999) Synergistic effect of histone hyperacetylation and DNA demethylation in the reactivation of the FMR1 gene. Hum Mol Genet 8:2317–2323 - PubMed
-
- Coffee B, Zhang F, Warren ST, Reines D (1999) Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat Genet 22:98–101 - PubMed
-
- Devys D, Biancalana V, Rousseau F, Boué J, Mandel JL, Oberlé I (1992) Analysis of full fragile X mutations in fetal tissues and monozygotic twins indicate that abnormal methylation and somatic heterogeneity are established early in development. Am J Med Genet 43:208–216 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

