Characterization of VPI pathogenicity island and CTXphi prophage in environmental strains of Vibrio cholerae
- PMID: 11466276
- PMCID: PMC99527
- DOI: 10.1128/JB.183.16.4737-4746.2001
Characterization of VPI pathogenicity island and CTXphi prophage in environmental strains of Vibrio cholerae
Abstract
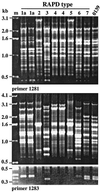
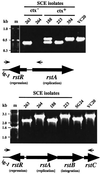
Environmental isolates of Vibrio cholerae of eight randomly amplified polymorphic DNA (RAPD) fingerprint types from Calcutta, India, that were unusual in containing toxin-coregulated pilus or cholera toxin genes but not O1 or O139 antigens of epidemic strains were studied by PCR and sequencing to gain insights into V. cholerae evolution. We found that each isolate contained a variant form of the VPI pathogenicity island. Distinguishing features included (i) four new alleles of tcpF (which encodes secreted virulence protein; its exact function is unknown), 20 to 70% divergent (at the protein level) from each other and canonical tcpF; (ii) a new allele of toxT (virulence regulatory gene), 36% divergent (at the protein level) in its 5' half and nearly identical in its 3' half to canonical toxT; (iii) a new tcpA (pilin) gene; and (iv) four variant forms of a regulatory sequence upstream of toxT. Also found were transpositions of an IS903-related element and function-unknown genes to sites in VPI. Cholera toxin (ctx) genes were found in isolates of two RAPD types, in each case embedded in CTXphi-like prophages. Fragments that are inferred to contain only putative repressor, replication, and integration genes were present in two other RAPD types. New possible prophage repressor and replication genes were also identified. Our results show marked genetic diversity in the virulence-associated gene clusters found in some nonepidemic V. cholerae strains, suggest that some of these genes contribute to fitness in nature, and emphasize the potential importance of interstrain gene exchange in the evolution of this species.
Figures





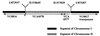

References
-
- Basu A, Mukhopadhyay A K, Sharma C, Jyot J, Gupta N, Ghosh A, Bhattacharya S K, Takeda Y, Faruque A S G, Albert M J, Nair G B. Heterogeneity in the organization of the CTX genetic element in strains of Vibrio choleraeO139 Bengal isolated from Calcutta, India and Dhaka, Bangladesh and its possible link to the dissimilar incidence of O139 cholera in the two locales. Microb Pathog. 1998;24:175–183. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

