Molecular characterization and gene content of breakpoint boundaries in patients with neurofibromatosis type 1 with 17q11.2 microdeletions
- PMID: 11468690
- PMCID: PMC1235482
- DOI: 10.1086/323043
Molecular characterization and gene content of breakpoint boundaries in patients with neurofibromatosis type 1 with 17q11.2 microdeletions
Abstract
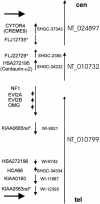
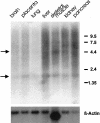
Homologous recombination between poorly characterized regions flanking the NF1 locus causes the constitutional loss of approximately 1.5 Mb from 17q11.2 covering > or =11 genes in 5%-20% of patients with neurofibromatosis type 1 (NF1). To elucidate the extent of microheterogeneity at the deletion boundaries, we used single-copy DNA fragments from the extreme ends of the deleted segment to perform FISH on metaphase chromosomes from eight patients with NF1 who had large deletions. In six patients, these probes were deleted, suggesting that breakage and fusions occurred within the adjacent highly homologous sequences. Reexamination of the deleted region revealed two novel functional genes FLJ12735 (AK022797) and KIAA0653-related (WI-12393 and AJ314647), the latter of which is located closest to the distal boundary and is partially duplicated. We defined the complete reading frames for these genes and two expressed-sequence tag (EST) clusters that were reported elsewhere and are associated with the markers SHGC-2390 and WI-9521. Hybrid cell lines carrying only the deleted chromosome 17 were generated from two patients and used to identify the fusion sequences by junction-specific PCRs. The proximal breakpoints were found between positions 125279 and 125479 in one patient and within 4 kb of position 143000 on BAC R-271K11 (AC005562) in three patients, and the distal breakpoints were found at the precise homologous position on R-640N20 (AC023278). The interstitial 17q11.2 microdeletion arises from unequal crossover between two highly homologous WI-12393-derived 60-kb duplicons separated by approximately 1.5 Mb. Since patients with the NF1 large-deletion syndrome have a significantly increased risk of neurofibroma development and mental retardation, hemizygosity for genes from the deleted region around the neurofibromin locus (CYTOR4, FLJ12735, FLJ22729, HSA272195 (centaurin-alpha2), NF1, OMGP, EVI2A, EVI2B, WI-9521, HSA272196, HCA66, KIAA0160, and WI-12393) may contribute to the severe phenotype of these patients.
Figures



References
Electronic-Database Information
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank (for IMAGE clone y174c12)
-
- Human Genome, http://www.ncbi.nlm.nih.gov/genome/guide/human (for draft sequences of the human genome)
-
- Institute for Genome Research, http://www.tigr.org (for draft sequences of the human genome)
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for NF1 [MIM 162200]) - PubMed
References
-
- Ars E, Serra E, Garcia J, Kruyer H, Gaona A, Lazaro C, Estivill X (2000) Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum Mol Genet 9:237–247 - PubMed
-
- Boerkoel CF, Inoue K, Reiter LT, Warner LE, Lupski JR (1999) Molecular mechanisms for CMT1A duplication and HNPP deletion. Ann N Y Acad Sci 883:22–35 - PubMed
-
- Cnossen MH, van der Est MN, Breuning MH, van Asperen CJ, Breslau-Siderius EJ, van der Ploeg AT, de Goede-Bolder A, van den Ouweland AM, Halley DJ, Niermeijer MF (1997) Deletions spanning the neurofibromatosis type 1 gene: implications for genotype-phenotype correlations in neurofibromatosis type 1? Hum Mutat 9:458–464 - PubMed
-
- Colman SD, Williams CA, Wallace MR (1995) Benign neurofibromas in type 1 neurofibromatosis (NF1) show somatic deletions of the NF1 gene. Nat Genet 11:90–92 - PubMed
-
- Dorschner MO, Sybert VP, Weaver M, Pletcher BA, Stephens K (2000) NF1 microdeletion breakpoints are clustered at flanking repetitive sequences. Hum Mol Genet 9:35–46 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

