mua-3, a gene required for mechanical tissue integrity in Caenorhabditis elegans, encodes a novel transmembrane protein of epithelial attachment complexes
- PMID: 11470828
- PMCID: PMC2150771
- DOI: 10.1083/jcb.200103035
mua-3, a gene required for mechanical tissue integrity in Caenorhabditis elegans, encodes a novel transmembrane protein of epithelial attachment complexes
Abstract
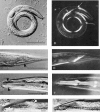
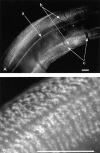
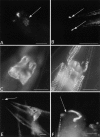
Normal locomotion of the nematode Caenorhabditis elegans requires transmission of contractile force through a series of mechanical linkages from the myofibrillar lattice of the body wall muscles, across an intervening extracellular matrix and epithelium (the hypodermis) to the cuticle. Mutations in mua-3 cause a separation of the hypodermis from the cuticle, suggesting this gene is required for maintaining hypodermal-cuticle attachment as the animal grows in size postembryonically. mua-3 encodes a predicted 3,767 amino acid protein with a large extracellular domain, a single transmembrane helix, and a smaller cytoplasmic domain. The extracellular domain contains four distinct protein modules: 5 low density lipoprotein type A, 52 epidermal growth factor, 1 von Willebrand factor A, and 2 sea urchin-enterokinase-agrin modules. MUA-3 localizes to the hypodermal hemidesmosomes and to other sites of mechanically robust transepithelial attachments, including the rectum, vulva, mechanosensory neurons, and excretory duct/pore. In addition, it is shown that MUA-3 colocalizes with cytoplasmic intermediate filaments (IFs) at these sites. Thus, MUA-3 appears to be a protein that links the IF cytoskeleton of nematode epithelia to the cuticle at sites of mechanical stress.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

