Folding and signaling share the same pathway in a photoreceptor
- PMID: 11470891
- PMCID: PMC55373
- DOI: 10.1073/pnas.111153598
Folding and signaling share the same pathway in a photoreceptor
Abstract
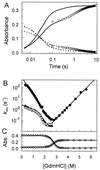
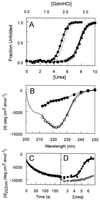
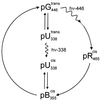
The photoreceptor photoactive yellow protein (PYP) was used as a model system to study receptor activation and protein folding. Refolding was studied by stopped-flow absorbance spectroscopy for PYP with either a trans or a cis chromophore. Chromophore trans to cis isomerization, the mechanism of light detection by PYP, greatly affects the protein folding process. When the cis chromophore is present, refolding from the unfolded state proceeds through the putative signaling state of PYP as an on-pathway intermediate. In addition, moderate denaturant concentrations result in the specific unfolding of the signaling state of PYP. Thus, the signaling state is common to the pathways of folding and signaling. This result provides an avenue for the study of protein folding. We demonstrate how this approach can be used to establish whether a folding intermediate is on-pathway or off-pathway. The results also reveal transient partial unfolding as a molecular mechanism for signaling.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

