Distribution of archaea in a black smoker chimney structure
- PMID: 11472939
- PMCID: PMC93063
- DOI: 10.1128/AEM.67.8.3618-3629.2001
Distribution of archaea in a black smoker chimney structure
Abstract
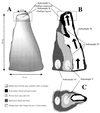
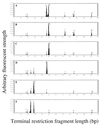
Archaeal community structures in microhabitats in a deep-sea hydrothermal vent chimney structure were evaluated through the combined use of culture-independent molecular analyses and enrichment culture methods. A black smoker chimney was obtained from the PACMANUS site in the Manus Basin near Papua New Guinea, and subsamples were obtained from vertical and horizontal sections. The elemental composition of the chimney was analyzed in different subsamples by scanning electron microscopy and energy-dispersive X-ray spectroscopy, indicating that zinc and sulfur were major components while an increased amount of elemental oxygen in exterior materials represented the presence of oxidized materials on the outer surface of the chimney. Terminal restriction fragment length polymorphism analysis revealed that a shift in archaeal ribotype structure occurred in the chimney structure. Through sequencing of ribosomal DNA (rDNA) clones from archaeal rDNA clone libraries, it was demonstrated that the archaeal communities in the chimney structure consisted for the most part of hyperthermophilic members and extreme halophiles and that the distribution of such extremophiles in different microhabitats of the chimney varied. The results of the culture-dependent analysis supported in part the view that changes in archaeal community structures in these microhabitats are associated with the geochemical and physical dynamics in the black smoker chimney.
Figures




References
-
- Bakken L R, Olsen R A. DNA content of soil bacteria of different cell size. Soil Biol Biochem. 1989;21:789–793.
-
- Baross J A. Isolation, growth, and maintenance of hyperthermophiles. In: Robb F T, Place A R, Sowers K R, Schreier H J, DasSarma S, Fleischmann E M, editors. Archaea: a laboratory manual: halophiles. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 15–24.
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

