Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid
- PMID: 11481432
- PMCID: PMC55547
- DOI: 10.1073/pnas.161294798
Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid
Abstract
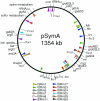
The symbiotic nitrogen-fixing soil bacterium Sinorhizobium meliloti contains three replicons: pSymA, pSymB, and the chromosome. We report here the complete 1,354,226-nt sequence of pSymA. In addition to a large fraction of the genes known to be specifically involved in symbiosis, pSymA contains genes likely to be involved in nitrogen and carbon metabolism, transport, stress, and resistance responses, and other functions that give S. meliloti an advantage in its specialized niche.
Figures
References
-
- Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, Watanabe A, Idesawa K, Ishikawa A, Kawashima K, et al. DNA Res. 2000;7:331–338. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

