The Calpha ---H...O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions
- PMID: 11481472
- PMCID: PMC55372
- DOI: 10.1073/pnas.161280798
The Calpha ---H...O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions
Abstract
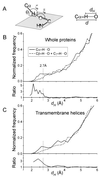
The Calpha---H...O hydrogen bond has been given little attention as a determinant of transmembrane helix association. Stimulated by recent calculations suggesting that such bonds can be much stronger than has been supposed, we have analyzed 11 known membrane protein structures and found that apparent carbon alpha hydrogen bonds cluster frequently at glycine-, serine-, and threonine-rich packing interfaces between transmembrane helices. Parallel right-handed helix-helix interactions appear to favor Calpha---H...O bond formation. In particular, Calpha---H...O interactions are frequent between helices having the structural motif of the glycophorin A dimer and the GxxxG pair. We suggest that Calpha---H...O hydrogen bonds are important determinants of stability and, depending on packing, specificity in membrane protein folding.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

