Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene
- PMID: 11483500
- PMCID: PMC149149
- DOI: 10.1093/emboj/20.15.3967
Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene
Abstract
A group of specialized genes has been defined to govern the molecular mechanisms controlling the circadian clock in mammals. Their expression and the interactions among their products dictate circadian rhythmicity. Three genes homologous to Drosophila period exist in the mouse and are thought to be major players in the biological clock. Here we present the generation of mice in which the founding member of the family, Per1, has been inactivated by homologous recombination. These mice present rhythmicity in locomotor activity, but with a period almost 1 h shorter than wild-type littermates. Moreover, the expression of clock genes in peripheral tissues appears to be delayed in Per1 mutant animals. Importantly, light-induced phase shifting appears conserved. The oscillatory expression of clock genes and the induction of immediate-early genes in response to light in the master clock structure, the suprachiasmatic nucleus, are unaffected. Altogether, these data demonstrate that Per1 plays a distinct role within the Per family, as it may be involved predominantly in peripheral clocks and/or in the output pathways of the circadian clock.
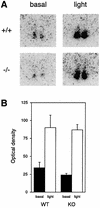
Figures





References
-
- Albrecht U., Sun,Z.S., Eichele,G. and Lee,C.C. (1997) A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell, 91, 1055–1064. - PubMed
-
- Balsalobre A., Damiola,F. and Schibler,U. (1998) A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell, 93, 929–937. - PubMed
-
- Brown S.A. and Schibler,U. (1999) The ins and outs of circadian timekeeping. Curr. Opin. Genet. Dev., 9, 588–594. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

