An essential nuclear envelope integral membrane protein, Brr6p, required for nuclear transport
- PMID: 11483521
- PMCID: PMC149179
- DOI: 10.1093/emboj/20.15.4183
An essential nuclear envelope integral membrane protein, Brr6p, required for nuclear transport
Abstract
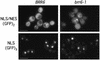
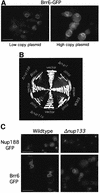
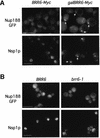
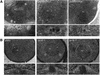
Despite rapid advances in our understanding of the function of the nuclear pore complex in nuclear transport, little is known about the role the nuclear envelope itself may play in this critical process. A small number of integral membrane proteins specific to the envelope have been identified in budding yeast, however, none has been reported to affect transport. We have identified an essential gene, BRR6, whose product, Brr6p, behaves like a nuclear envelope integral membrane protein. Notably, the brr6-1 mutant specifically affects transport of mRNA and a protein reporter containing a nuclear export signal. In addition, Brr6p depletion alters nucleoporin distribution and nuclear envelope morphology, suggesting that the protein is required for the spatial organization of nuclear pores. BRR6 interacts genetically with a subset of nucleoporins, and Brr6-green fluorescent protein (GFP) localizes in a punctate nuclear rim pattern, suggesting location at or near the nuclear pore. However, Brr6-GFP fails to redistribute in a (Delta)nup133 mutant, distinguishing Brr6p from known proteins of the pore membrane domain. We hypothesize that Brr6p is located adjacent to the nuclear pore and interacts functionally with the pore and transport machinery.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

