The kink-turn: a new RNA secondary structure motif
- PMID: 11483524
- PMCID: PMC149158
- DOI: 10.1093/emboj/20.15.4214
The kink-turn: a new RNA secondary structure motif
Abstract
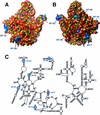
Analysis of the Haloarcula marismortui large ribosomal subunit has revealed a common RNA structure that we call the kink-turn, or K-turn. The six K-turns in H.marismortui 23S rRNA superimpose with an r.m.s.d. of 1.7 A. There are two K-turns in the structure of Thermus thermophilus 16S rRNA, and the structures of U4 snRNA and L30e mRNA fragments form K-turns. The structure has a kink in the phosphodiester backbone that causes a sharp turn in the RNA helix. Its asymmetric internal loop is flanked by C-G base pairs on one side and sheared G-A base pairs on the other, with an A-minor interaction between these two helical stems. A derived consensus secondary structure for the K-turn includes 10 consensus nucleotides out of 15, and predicts its presence in the 5'-UTR of L10 mRNA, helix 78 in Escherichia coli 23S rRNA and human RNase MRP. Five K-turns in 23S rRNA interact with nine proteins. While the observed K-turns interact with proteins of unrelated structures in different ways, they interact with L7Ae and two homologous proteins in the same way.
Figures








References
-
- Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. - PubMed
-
- Beckmann R., Bubeck,D., Grassucci,R., Penczek,P., Verschoor,A., Blobel,G. and Frank,J. (1997) Alignment of conduits for the nascent polypeptide chain in the ribosome–Sec61 complex. Science, 278, 2123–2126. - PubMed
-
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
-
- Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. - PubMed
-
- Cate J.H., Yusupov,M.M., Yusupova,G.Z., Earnest,T.N. and Noller,H.F. (1999) X-ray crystal structures of 70S ribosome functional complexes. Science, 285, 2095–2104. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

