Non-canonical mechanism for translational control in bacteria: synthesis of ribosomal protein S1
- PMID: 11483525
- PMCID: PMC149162
- DOI: 10.1093/emboj/20.15.4222
Non-canonical mechanism for translational control in bacteria: synthesis of ribosomal protein S1
Abstract
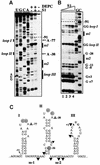
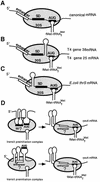
Translation initiation region (TIR) of the rpsA mRNA encoding ribosomal protein S1 is one of the most efficient in Escherichia coli despite the absence of a canonical Shine-Dalgarno-element. Its high efficiency is under strong negative autogenous control, a puzzling phenomenon as S1 has no strict sequence specificity. To define sequence and structural elements responsible for translational efficiency and autoregulation of the rpsA mRNA, a series of rpsA'-'lacZ chromosomal fusions bearing various mutations in the rpsA TIR was created and tested for beta-galactosidase activity in the absence and presence of excess S1. These in vivo results, as well as data obtained by in vitro techniques and phylogenetic comparison, allow us to propose a model for the structural and functional organization of the rpsA TIR specific for proteobacteria related to E.coli. According to the model, the high efficiency of translation initiation is provided by a specific fold of the rpsA leader forming a non-contiguous ribosome entry site, which is destroyed upon binding of free S1 when it acts as an autogenous repressor.
Figures






References
-
- Bycroft M., Hubbard,T.J., Proctor,M., Freund,S.M. and Murzin,A.G. (1997) The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell, 88, 235–242. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

