Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation
- PMID: 11483526
- PMCID: PMC149147
- DOI: 10.1093/emboj/20.15.4233
Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation
Abstract
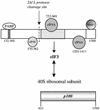
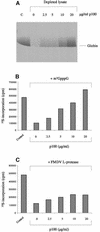
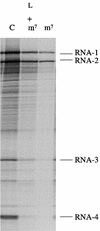
Picornavirus proteases cleave translation initiation factor eIF4G into a C-terminal two-thirds fragment (hereafter named p100) and an N-terminal one-third fragment, which interacts with the cap-binding factor eIF4E. As the timing of this cleavage correlates broadly with the shut-off of host cell protein synthesis in infected cells, a very widespread presumption has been that p100 cannot support capped mRNA translation. Through the use of an eIF4G-depleted reticulocyte lysate system, we show that this presumption is incorrect. Moreover, recombinant p100 can also reverse the inhibition of capped mRNA translation caused either by m7GpppG cap analogue, by 4E-BP1, which sequesters eIF4E and thus blocks its association with eIF4G, or by cleavage of endogenous eIF4G by picornavirus proteases. The concentration of p100 required for maximum translation of capped mRNAs is approximately 4-fold higher than the endogenous eIF4G concentration in reticulocyte lysates. Our results imply that picornavirus-induced shut-off is not due to an intrinsic inability of p100 to support capped mRNA translation, but to the viral RNA outcompeting host cell mRNA for the limiting concentration of p100.
Figures





References
-
- Ahlquist P., Luckow,V. and Kaesberg,P. (1981) Complete nucleotide sequence of Brome Mosaic Virus RNA 3. J. Mol. Biol., 153, 23–38. - PubMed
-
- Ahlquist P., Dasgupta,R. and Kaesberg,P. (1984) Nucleotide sequence of the Brome Mosaic Virus genome and its implications for viral replication. J. Mol. Biol., 172, 369–383. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

