Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G
- PMID: 11483729
- PMCID: PMC115028
- DOI: 10.1128/jvi.75.17.7854-7863.2001
Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G
Abstract
The question of whether translation initiation factor eIF4E and the complete eIF4G polypeptide are required for initiation dependent on the IRES (internal ribosome entry site) of hepatitis A virus (HAV) has been examined using in vitro translation in standard and eIF4G-depleted rabbit reticulocyte lysates. In agreement with previous publications, the HAV IRES is unique among all picornavirus IRESs in that it was inhibited if translation initiation factor eIF4G was cleaved by foot-and-mouth disease L-proteases. In addition, the HAV IRES was inhibited by addition of eIF4E-binding protein 1, which binds tightly to eIF4E and sequesters it, thus preventing its association with eIF4G. The HAV IRES was also inhibited by addition of m(7)GpppG cap analogue, irrespective of whether the RNA tested was capped or not. Thus, initiation on the HAV IRES requires that eIF4E be associated with eIF4G and that the cap-binding pocket of eIF4E be empty and unoccupied. This suggests two alternative models: (i) initiation requires a direct interaction between an internal site in the IRES and eIF4E/4G, an interaction which involves the cap-binding pocket of eIF4E in addition to any direct eIF4G-RNA interactions; or (ii) it requires eIF4G in a particular conformation which can be attained only if eIF4E is bound to it, with the cap-binding pocket of the eIF4E unoccupied.
Figures

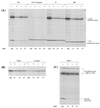
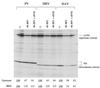




References
-
- Belsham G J, Jackson R J. Translation initiation on picornavirus RNA. In: Sonenberg N, Hershey J W B, Mathews M B, editors. Translational control of gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. pp. 869–900.
-
- Borman A, Jackson R J. Initiation of translation of human rhinovirus RNA: mapping the internal ribosome entry site. Virology. 1992;188:685–696. - PubMed
-
- Borman A M, Kean K M. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology. 1997;237:129–136. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

