Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells
- PMID: 11483742
- PMCID: PMC115041
- DOI: 10.1128/jvi.75.17.7973-7986.2001
Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells
Abstract
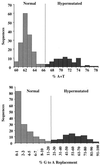
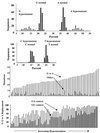
G-to-A hypermutation has been sporadically observed in human immunodeficiency virus type 1 (HIV-1) proviral sequences from patient peripheral blood mononuclear cells (PBMC) and virus cultures but has not been systematically evaluated. PCR primers matched to normal and hypermutated sequences were used in conjunction with an agarose gel electrophoresis system incorporating an AT-binding dye to visualize, separate, clone, and sequence hypermutated and normal sequences in the 297-bp HIV-1 protease gene amplified from patient PBMC. Among 53 patients, including individuals infected with subtypes A through D and at different clinical stages, at least 43% of patients harbored abundant hypermutated, along with normal, protease genes. In 70 hypermutated sequences, saturation of G residues in the GA or GG dinucleotide context ranged from 20 to 94%. Levels of other mutants were not elevated, and G-to-A replacement was entirely restricted to GA or GG, and not GC or GT, dinucleotides. Sixty-nine of 70 hypermutated and 3 of 149 normal sequences had in-frame stop codons. To investigate the conditions under which hypermutation occurs in cell cultures, purified CD4(+) T cells from normal donors were infected with cloned NL4-3 virus stocks at various times before and after phytohemagglutinin (PHA) activation. Hypermutation was pronounced when HIV-1 infection occurred simultaneously with, or a few hours after, PHA activation, but after 12 h or more after PHA activation, most HIV-1 sequences were normal. Hypermutated sequences generated in culture corresponded exactly in all parameters to those obtained from patient PBMC. Near-simultaneous activation and infection of CD4(+) T cells may represent a window of susceptibility where the informational content of HIV-1 sequences is lost due to hypermutation.
Figures





References
-
- Bebenek K, Abbotts J, Roberts J D, Wilson S H, Kunkel T A. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989;264:16948–16956. - PubMed
-
- Bianchi V, Borella S, Rampazzo C, Ferraro P, Calderazzo F, Bianchi L C, Skog S, Reichard P. Cell cycle-dependent metabolism of pyrimidine deoxynucleoside triphosphates in CEM cells. J Biol Chem. 1997;272:16118–16124. - PubMed
-
- Borman A M, Quillent C, Charneau P, Kean K M, Clavel F. A highly defective HIV-1 group O provirus: evidence for the role of local sequence determinants in G→A hypermutation during negative-strand viral DNA synthesis. Virology. 1995;208:601–609. - PubMed
-
- Bray G, Brent T P. Deoxyribonucleoside 5′-triphosphate pool fluctuations during the mammalian cell cycle. Biochim Biophys Acta. 1972;269:184–191. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

