Host switching in Lyssavirus history from the Chiroptera to the Carnivora orders
- PMID: 11483755
- PMCID: PMC115054
- DOI: 10.1128/jvi.75.17.8096-8104.2001
Host switching in Lyssavirus history from the Chiroptera to the Carnivora orders
Abstract
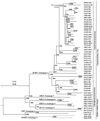
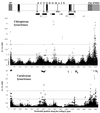
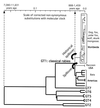
Lyssaviruses are unsegmented RNA viruses causing rabies. Their vectors belong to the Carnivora and Chiroptera orders. We studied 36 carnivoran and 17 chiropteran lyssaviruses representing the main genotypes and variants. We compared their genes encoding the surface glycoprotein, which is responsible for receptor recognition and membrane fusion. The glycoprotein is the main protecting antigen and bears virulence determinants. Point mutation is the main force in lyssavirus evolution, as Sawyer's test and phylogenetic analysis showed no evidence of recombination. Tests of neutrality indicated a neutral model of evolution, also supported by globally high ratios of synonymous substitutions (d(S)) to nonsynonymous substitutions (d(N)) (>7). Relative-rate tests suggested similar rates of evolution for all lyssavirus lineages. Therefore, the absence of recombination and similar evolutionary rates make phylogeny-based conclusions reliable. Phylogenetic reconstruction strongly supported the hypothesis that host switching occurred in the history of lyssaviruses. Indeed, lyssaviruses evolved in chiropters long before the emergence of carnivoran rabies, very likely following spillovers from bats. Using dated isolates, the average rate of evolution was estimated to be roughly 4.3 x 10(-4) d(S)/site/year. Consequently, the emergence of carnivoran rabies from chiropteran lyssaviruses was determined to have occurred 888 to 1,459 years ago. Glycoprotein segments accumulating more d(N) than d(S) were distinctly detected in carnivoran and chiropteran lyssaviruses. They may have contributed to the adaptation of the virus to the two distinct mammal orders. In carnivoran lyssaviruses they overlapped the main antigenic sites, II and III, whereas in chiropteran lyssaviruses they were located in regions of unknown functions.
Figures
References
-
- Aaziz R, Tepfer M. Recombination in RNA viruses and in virus-resistant transgenic plants. J Gen Virol. 1999;80:1339–1346. - PubMed
-
- Aitken T H, Kowalski R W, Beaty B J, Buckley S M, Wright J D, Shope R E, Miller B R. Arthropod studies with rabies-related Mokola virus. Am J Trop Med Hyg. 1984;33:945–952. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

