RNAs extracted from herpes simplex virus 1 virions: apparent selectivity of viral but not cellular RNAs packaged in virions
- PMID: 11483756
- PMCID: PMC115055
- DOI: 10.1128/jvi.75.17.8105-8116.2001
RNAs extracted from herpes simplex virus 1 virions: apparent selectivity of viral but not cellular RNAs packaged in virions
Abstract
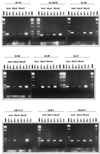
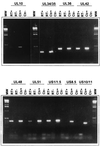
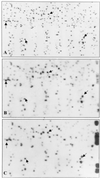
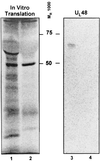
Following the lead of recent studies on the presence of RNA in virions of human cytomegalovirus, we investigated the presence and identity of RNAs from purified virions of herpes simple virus 1. To facilitate these studies, we designed primers for all known open reading frames (ORFs) and also constructed cDNA arrays containing probes designed to detect all known transcripts. In the first series of experiments, labeled DNA made by reverse transcription of poly(A)(+) RNA extracted from infected HEp-2 or rabbit skin cells hybridized to all but two of the probes in the cDNA array. A similar analysis of the RNA extracted from purified extracellular virions derived from infected HEp-2 cells hybridized to probes representing 24 of the ORFs. In the second series of analyses, we reverse transcribed and amplified by PCR RNAs from purified intracellular or extracellular virions derived from infected HEp-2 or Vero cell lines. The positive RNAs were retested by PCR with and without prior reverse transcription to ensure that the samples tested were free of contaminating DNA. The results were as follows. (i) Only a fraction of viral ORF transcripts were represented in virion RNA, and only nine RNAs (U(L)10, U(L)34/U(L)35, U(L)36, U(L)42, U(L)48, U(L)51, U(S)1/U(S)1.5, U(S)8.5, and U(S)10/U(S)11) were positive in all RT PCR assays. Of these, seven were positive by hybridization to cDNA arrays. (ii) RNA extracted from cells infected with a mutant virus lacking the U(S)8 to U(S)12 genes yielded results similar to those described above, indicating that U(S)11, a known RNA binding protein, does not play a role in packaging RNA in virions. (iii) Cellular RNAs detected in virions were representative of the abundant cellular RNAs. Last, RNA extracted from virions was translated in vitro and the translation products were reacted with antibody to alphaTIF (VIP16). The immune precipitate contained a labeled protein with the apparent molecular weight of alphaTIF, indicating that at least one mRNA packaged in virions was intact and capable of being translated. The basis for the apparent selectivity in the packaging of the viral RNAs packaged in virions is unknown.
Figures


Similar articles
-
Of the three tegument proteins that package mRNA in herpes simplex virions, one (VP22) transports the mRNA to uninfected cells for expression prior to viral infection.Proc Natl Acad Sci U S A. 2002 Jun 11;99(12):8318-23. doi: 10.1073/pnas.122231699. Proc Natl Acad Sci U S A. 2002. PMID: 12060774 Free PMC article.
-
Viral RNAs detected in virions of porcine adenovirus type 3.Virology. 2004 Apr 10;321(2):372-82. doi: 10.1016/j.virol.2003.12.025. Virology. 2004. PMID: 15051396
-
Herpes simplex virus DNA cleavage and packaging: association of multiple forms of U(L)15-encoded proteins with B capsids requires at least the U(L)6, U(L)17, and U(L)28 genes.J Virol. 1998 Apr;72(4):3045-50. doi: 10.1128/JVI.72.4.3045-3050.1998. J Virol. 1998. PMID: 9525627 Free PMC article.
-
[Analysis on primate lentivirus genome dimerization in virion].Uirusu. 2005 Jun;55(1):153-60. doi: 10.2222/jsv.55.153. Uirusu. 2005. PMID: 16308542 Review. Japanese.
-
Purification Methods and the Presence of RNA in Virus Particles and Extracellular Vesicles.Viruses. 2020 Aug 21;12(9):917. doi: 10.3390/v12090917. Viruses. 2020. PMID: 32825599 Free PMC article. Review.
Cited by
-
Recognition of herpesviruses by the innate immune system.Nat Rev Immunol. 2011 Feb;11(2):143-54. doi: 10.1038/nri2937. Nat Rev Immunol. 2011. PMID: 21267015 Free PMC article. Review.
-
Resistance of mRNA translation to acute endoplasmic reticulum stress-inducing agents in herpes simplex virus type 1-infected cells requires multiple virus-encoded functions.J Virol. 2006 Aug;80(15):7354-63. doi: 10.1128/JVI.00479-06. J Virol. 2006. PMID: 16840316 Free PMC article.
-
RNAs in the virion of Kaposi's sarcoma-associated herpesvirus.J Virol. 2005 Aug;79(16):10138-46. doi: 10.1128/JVI.79.16.10138-10146.2005. J Virol. 2005. PMID: 16051806 Free PMC article.
-
Adventitious Virus Detection in Cells by High-Throughput Sequencing of Newly Synthesized RNAs: Unambiguous Differentiation of Cell Infection from Carryover of Viral Nucleic Acids.mSphere. 2019 Jun 5;4(3):e00298-19. doi: 10.1128/mSphere.00298-19. mSphere. 2019. PMID: 31167947 Free PMC article.
-
A virus-like particle-based Epstein-Barr virus vaccine.J Virol. 2011 Dec;85(24):13105-13. doi: 10.1128/JVI.05598-11. Epub 2011 Oct 12. J Virol. 2011. PMID: 21994444 Free PMC article.
References
-
- Brensnahan W A, Shenk T. A subset of viral transcripts packaged within human cytomegalovirus particles. Science. 2000;288:2373–2376. - PubMed
-
- Ejercito P M, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J Gen Virol. 1968;2:357–364. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

