Use of whole genome sequence data to infer baculovirus phylogeny
- PMID: 11483757
- PMCID: PMC115056
- DOI: 10.1128/jvi.75.17.8117-8126.2001
Use of whole genome sequence data to infer baculovirus phylogeny
Abstract
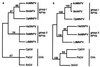
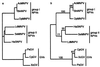
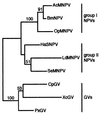
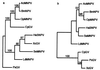
Several phylogenetic methods based on whole genome sequence data were evaluated using data from nine complete baculovirus genomes. The utility of three independent character sets was assessed. The first data set comprised the sequences of the 63 genes common to these viruses. The second set of characters was based on gene order, and phylogenies were inferred using both breakpoint distance analysis and a novel method developed here, termed neighbor pair analysis. The third set recorded gene content by scoring gene presence or absence in each genome. All three data sets yielded phylogenies supporting the separation of the Nucleopolyhedrovirus (NPV) and Granulovirus (GV) genera, the division of the NPVs into groups I and II, and species relationships within group I NPVs. Generation of phylogenies based on the combined sequences of all 63 shared genes proved to be the most effective approach to resolving the relationships among the group II NPVs and the GVs. The history of gene acquisitions and losses that have accompanied baculovirus diversification was visualized by mapping the gene content data onto the phylogenetic tree. This analysis highlighted the fluid nature of baculovirus genomes, with evidence of frequent genome rearrangements and multiple gene content changes during their evolution. Of more than 416 genes identified in the genomes analyzed, only 63 are present in all nine genomes, and 200 genes are found only in a single genome. Despite this fluidity, the whole genome-based methods we describe are sufficiently powerful to recover the underlying phylogeny of the viruses.
Figures
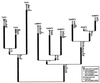
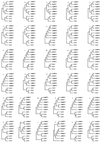
References
-
- Ahrens C H, Russell R L Q, Funk C J, Evans J T, Harwood S H, Rohrmann G F. The sequence of the Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus genome. Virology. 1997;229:381–399. - PubMed
-
- Ayres M D, Howard S C, Kuzio J, Lopez-Ferber M, Possee R D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. - PubMed
-
- Bideshi D K, Bigot Y, Federici B A. Molecular characterization and phylogenetic analysis of the Harrisina brillians granulovirus granulin gene. Arch Virol. 2000;145:1933–1945. - PubMed
-
- Blanchette M, Kunisawa T, Sankoff D. Gene order breakpoint evidence in animal mitochondrial phylogeny. J Mol Evol. 1999;49:193–203. - PubMed
-
- Blissard G W, Black B, Crook N, Keddie B A, Possee R, Rohrmann G, Theilmann D A, Volkman L. Seventh report of the international committee on taxonomy of viruses. In: Van Regenmortel H V, Bishop D H L, Van Regenmortel M H, Fauquet Claude M, editors. Virus taxonomy. San Diego, Calif: Academic Press; 2000. pp. 195–202.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

