Early polyadenylation signals of human papillomavirus type 31 negatively regulate capsid gene expression
- PMID: 11483760
- PMCID: PMC115059
- DOI: 10.1128/jvi.75.17.8147-8157.2001
Early polyadenylation signals of human papillomavirus type 31 negatively regulate capsid gene expression
Abstract
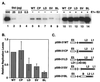
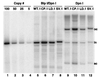
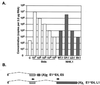
The L1 and L2 capsid genes of human papillomavirus type 31 (HPV-31) are expressed upon keratinocyte differentiation from a promoter located in the E7 open reading frame (ORF) of the early region. Late transcripts must therefore pass through and ignore the early polyadenylation sequences to use the downstream late AAUAAA element located at the end of the L1 ORF. To identify sequences which modulate downstream capsid gene expression, a variety of substitution mutations were introduced into the early polyadenylation signal and studied first in the context of polycistronic luciferase reporter constructs. Removal of the G/U-rich cleavage stimulation factor (CstF) binding sites and the degenerate cleavage and polyadenylation specificity factor binding sites, UAUAUA, had minimal effect on downstream expression as defined by luciferase activities. This is in contrast to the deletion of the HPV-31 early AAUAAA element, which resulted in a dramatic increase in downstream expression. Additional sequences within the first 800 bp of the L2 ORF were also found to negatively regulate capsid expression in luciferase assays. To determine how these mutations influence gene expression in the context of the complete HPV-31 genome, recombinant genomes were constructed that contained a substitution in the AAUAAA sequence, an inserted strong CstF binding site, an inserted simian virus 40 (SV40) late poly(A) signal, or a substitution of the 5'-most 800 nucleotides of the L2 ORF. Reductions in both transient and stable replication were observed with the recombinant genomes containing the strong CstF site or the late SV40 signal, suggesting that alterations in the strength of the upstream poly(A) signal influence expression of viral replication factors. Similarly, disruption of the L2 ORF resulted in a significant reduction in genome replication and an inability to be maintained stably. In contrast, genomes containing a substitution of the AAUAAA sequence had increased levels of transient and stable replication. Quantitation of late transcripts following keratinocyte differentiation in methylcellulose also showed a reduction in downstream capsid gene expression in lines containing genomes with the strong CstF site or the late SV40 signal mutations, while a significant increase in expression was detected in the lines with genomes lacking the AAUAAA sequence. These studies demonstrate that capsid gene expression in HPV-31 requires an inefficient early poly(A) signal which is defined primarily by the AAUAAA element as well as a major negative regulatory element located within the L2 ORF.
Figures





References
-
- Baker C C. Post-transcriptional regulation of papillomavirus gene expression., p. III-11–III-16. In: Myers G, Baker C, Munger K, Sverdrup F, McBride A, Bernard H-U, Meissner J, editors. Human papillomaviruses. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1997.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

